
Capybara microvirus Cap3_SP_316
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.71
Get precalculated fractions of proteins
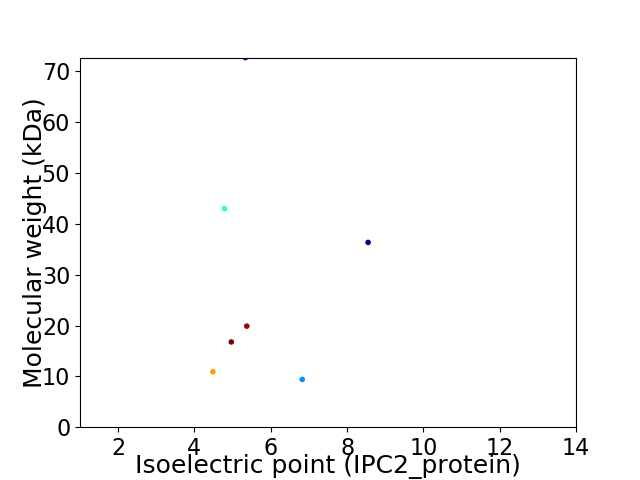
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
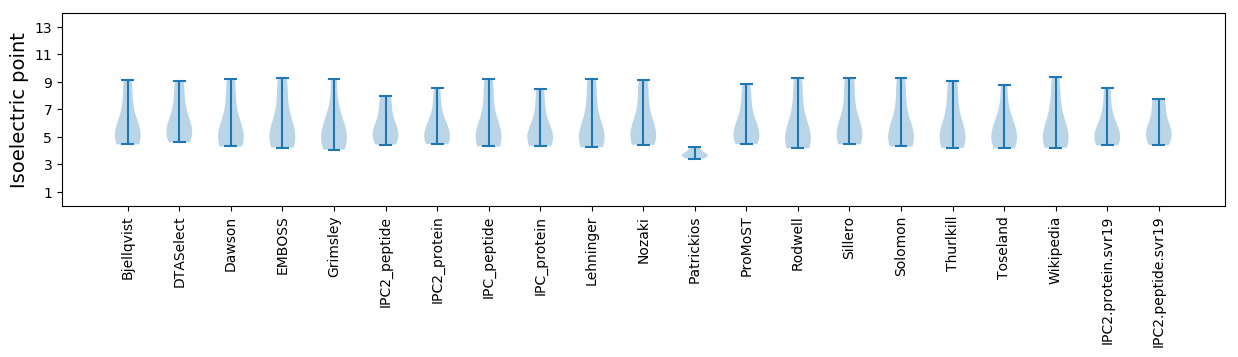
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVR9|A0A4V1FVR9_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_316 OX=2585426 PE=3 SV=1
MM1 pKa = 6.85TTDD4 pKa = 3.26KK5 pKa = 11.08LFYY8 pKa = 10.35IYY10 pKa = 9.74DD11 pKa = 3.9TVSDD15 pKa = 4.32LNLGYY20 pKa = 10.42RR21 pKa = 11.84FYY23 pKa = 10.01PTIGALIRR31 pKa = 11.84SEE33 pKa = 4.41APTISKK39 pKa = 9.19QFPNFEE45 pKa = 4.28TEE47 pKa = 4.03LKK49 pKa = 10.15IIEE52 pKa = 4.47IGSFVSDD59 pKa = 2.93SGYY62 pKa = 10.83DD63 pKa = 3.06ISSFNPLSVLDD74 pKa = 3.97IPLIHH79 pKa = 6.62SWDD82 pKa = 3.75EE83 pKa = 3.83YY84 pKa = 11.16SFQSSEE90 pKa = 4.18TKK92 pKa = 10.18FNKK95 pKa = 9.97
MM1 pKa = 6.85TTDD4 pKa = 3.26KK5 pKa = 11.08LFYY8 pKa = 10.35IYY10 pKa = 9.74DD11 pKa = 3.9TVSDD15 pKa = 4.32LNLGYY20 pKa = 10.42RR21 pKa = 11.84FYY23 pKa = 10.01PTIGALIRR31 pKa = 11.84SEE33 pKa = 4.41APTISKK39 pKa = 9.19QFPNFEE45 pKa = 4.28TEE47 pKa = 4.03LKK49 pKa = 10.15IIEE52 pKa = 4.47IGSFVSDD59 pKa = 2.93SGYY62 pKa = 10.83DD63 pKa = 3.06ISSFNPLSVLDD74 pKa = 3.97IPLIHH79 pKa = 6.62SWDD82 pKa = 3.75EE83 pKa = 3.83YY84 pKa = 11.16SFQSSEE90 pKa = 4.18TKK92 pKa = 10.18FNKK95 pKa = 9.97
Molecular weight: 10.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4Q1|A0A4P8W4Q1_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_316 OX=2585426 PE=4 SV=1
MM1 pKa = 7.63ICRR4 pKa = 11.84NPIDD8 pKa = 3.75SSNFKK13 pKa = 10.83YY14 pKa = 10.21PIYY17 pKa = 10.11DD18 pKa = 3.27ALGRR22 pKa = 11.84TVKK25 pKa = 10.33VPCNTCLACRR35 pKa = 11.84ANKK38 pKa = 9.3ILNYY42 pKa = 8.69SARR45 pKa = 11.84ISYY48 pKa = 9.83EE49 pKa = 3.61RR50 pKa = 11.84INNPEE55 pKa = 3.9CSFVTFTYY63 pKa = 10.73DD64 pKa = 4.43KK65 pKa = 11.46NHH67 pKa = 6.37LKK69 pKa = 10.96YY70 pKa = 10.9NFGFYY75 pKa = 10.76EE76 pKa = 4.19PTLKK80 pKa = 10.69RR81 pKa = 11.84SDD83 pKa = 3.1LHH85 pKa = 8.49KK86 pKa = 11.13YY87 pKa = 8.29IDD89 pKa = 3.89KK90 pKa = 10.54VRR92 pKa = 11.84HH93 pKa = 5.86LLPKK97 pKa = 10.14DD98 pKa = 3.44VKK100 pKa = 10.45FKK102 pKa = 11.14YY103 pKa = 9.49FASGQYY109 pKa = 10.09GDD111 pKa = 4.05RR112 pKa = 11.84FNRR115 pKa = 11.84PHH117 pKa = 5.7YY118 pKa = 9.7HH119 pKa = 5.55VLFIGLDD126 pKa = 3.43FVKK129 pKa = 10.51YY130 pKa = 10.26YY131 pKa = 11.18KK132 pKa = 10.42FFKK135 pKa = 10.62DD136 pKa = 3.07SWNLGSVDD144 pKa = 4.11VQPMLSGSVQYY155 pKa = 10.63VCSYY159 pKa = 10.59LHH161 pKa = 5.87HH162 pKa = 6.61QKK164 pKa = 10.66FGSDD168 pKa = 3.3SLDD171 pKa = 3.09SQFFDD176 pKa = 3.96NGLDD180 pKa = 3.62EE181 pKa = 5.46PFLSMSPGIGSDD193 pKa = 4.07FYY195 pKa = 10.97LAHH198 pKa = 7.02IFEE201 pKa = 5.45LLNGLPVFYY210 pKa = 10.16GSSSVQVPSYY220 pKa = 10.68YY221 pKa = 9.69RR222 pKa = 11.84RR223 pKa = 11.84KK224 pKa = 9.62YY225 pKa = 10.67LKK227 pKa = 10.5LSQFSLDD234 pKa = 3.88NIRR237 pKa = 11.84DD238 pKa = 3.84SLVLNRR244 pKa = 11.84KK245 pKa = 6.98YY246 pKa = 10.43HH247 pKa = 5.17YY248 pKa = 11.14NSMLEE253 pKa = 3.63AGYY256 pKa = 9.13TDD258 pKa = 3.86FGKK261 pKa = 10.09FVKK264 pKa = 9.66MKK266 pKa = 10.64SEE268 pKa = 3.89QQDD271 pKa = 3.52FQLINSLRR279 pKa = 11.84SRR281 pKa = 11.84GCPISQSYY289 pKa = 10.43LFGNTVYY296 pKa = 10.98SFDD299 pKa = 3.55KK300 pKa = 11.37GFINSLANSALKK312 pKa = 10.58AA313 pKa = 3.66
MM1 pKa = 7.63ICRR4 pKa = 11.84NPIDD8 pKa = 3.75SSNFKK13 pKa = 10.83YY14 pKa = 10.21PIYY17 pKa = 10.11DD18 pKa = 3.27ALGRR22 pKa = 11.84TVKK25 pKa = 10.33VPCNTCLACRR35 pKa = 11.84ANKK38 pKa = 9.3ILNYY42 pKa = 8.69SARR45 pKa = 11.84ISYY48 pKa = 9.83EE49 pKa = 3.61RR50 pKa = 11.84INNPEE55 pKa = 3.9CSFVTFTYY63 pKa = 10.73DD64 pKa = 4.43KK65 pKa = 11.46NHH67 pKa = 6.37LKK69 pKa = 10.96YY70 pKa = 10.9NFGFYY75 pKa = 10.76EE76 pKa = 4.19PTLKK80 pKa = 10.69RR81 pKa = 11.84SDD83 pKa = 3.1LHH85 pKa = 8.49KK86 pKa = 11.13YY87 pKa = 8.29IDD89 pKa = 3.89KK90 pKa = 10.54VRR92 pKa = 11.84HH93 pKa = 5.86LLPKK97 pKa = 10.14DD98 pKa = 3.44VKK100 pKa = 10.45FKK102 pKa = 11.14YY103 pKa = 9.49FASGQYY109 pKa = 10.09GDD111 pKa = 4.05RR112 pKa = 11.84FNRR115 pKa = 11.84PHH117 pKa = 5.7YY118 pKa = 9.7HH119 pKa = 5.55VLFIGLDD126 pKa = 3.43FVKK129 pKa = 10.51YY130 pKa = 10.26YY131 pKa = 11.18KK132 pKa = 10.42FFKK135 pKa = 10.62DD136 pKa = 3.07SWNLGSVDD144 pKa = 4.11VQPMLSGSVQYY155 pKa = 10.63VCSYY159 pKa = 10.59LHH161 pKa = 5.87HH162 pKa = 6.61QKK164 pKa = 10.66FGSDD168 pKa = 3.3SLDD171 pKa = 3.09SQFFDD176 pKa = 3.96NGLDD180 pKa = 3.62EE181 pKa = 5.46PFLSMSPGIGSDD193 pKa = 4.07FYY195 pKa = 10.97LAHH198 pKa = 7.02IFEE201 pKa = 5.45LLNGLPVFYY210 pKa = 10.16GSSSVQVPSYY220 pKa = 10.68YY221 pKa = 9.69RR222 pKa = 11.84RR223 pKa = 11.84KK224 pKa = 9.62YY225 pKa = 10.67LKK227 pKa = 10.5LSQFSLDD234 pKa = 3.88NIRR237 pKa = 11.84DD238 pKa = 3.84SLVLNRR244 pKa = 11.84KK245 pKa = 6.98YY246 pKa = 10.43HH247 pKa = 5.17YY248 pKa = 11.14NSMLEE253 pKa = 3.63AGYY256 pKa = 9.13TDD258 pKa = 3.86FGKK261 pKa = 10.09FVKK264 pKa = 9.66MKK266 pKa = 10.64SEE268 pKa = 3.89QQDD271 pKa = 3.52FQLINSLRR279 pKa = 11.84SRR281 pKa = 11.84GCPISQSYY289 pKa = 10.43LFGNTVYY296 pKa = 10.98SFDD299 pKa = 3.55KK300 pKa = 11.37GFINSLANSALKK312 pKa = 10.58AA313 pKa = 3.66
Molecular weight: 36.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1846 |
77 |
650 |
263.7 |
29.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.629 ± 0.463 | 1.138 ± 0.385 |
7.367 ± 0.616 | 3.575 ± 0.433 |
7.259 ± 0.542 | 5.525 ± 0.53 |
1.463 ± 0.333 | 6.013 ± 0.613 |
5.742 ± 0.7 | 9.751 ± 0.4 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.138 ± 0.143 | 7.638 ± 0.388 |
4.28 ± 0.882 | 3.359 ± 0.375 |
4.009 ± 0.21 | 13.868 ± 0.935 |
3.738 ± 0.647 | 4.388 ± 0.529 |
0.704 ± 0.162 | 5.417 ± 0.679 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
