
Krasilnikoviella flava
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Promicromonosporaceae; Krasilnikoviella
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
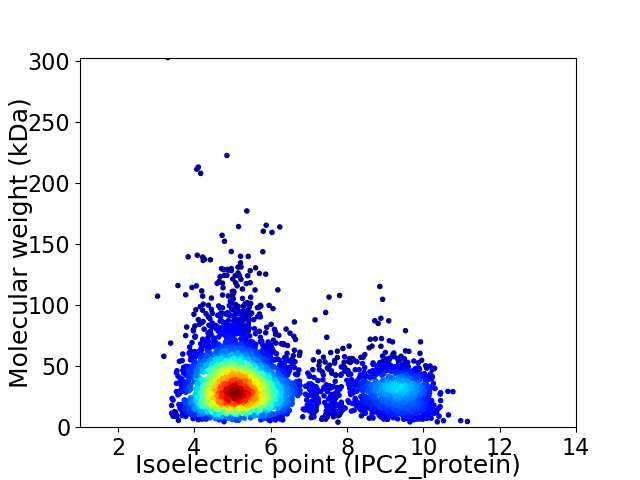
Virtual 2D-PAGE plot for 4296 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1T5JMS5|A0A1T5JMS5_9MICO Amidohydrolase OS=Krasilnikoviella flava OX=526729 GN=SAMN04324258_1562 PE=4 SV=1
MM1 pKa = 7.78RR2 pKa = 11.84RR3 pKa = 11.84GLRR6 pKa = 11.84RR7 pKa = 11.84AVAASAAGLAGLSLAACAPGGDD29 pKa = 4.32GAPADD34 pKa = 3.79NTSCTNEE41 pKa = 3.88IKK43 pKa = 11.11VPDD46 pKa = 3.93ATRR49 pKa = 11.84VTLWAWYY56 pKa = 8.02PAVEE60 pKa = 4.59QIVDD64 pKa = 3.73TFNAQHH70 pKa = 7.48DD71 pKa = 4.24DD72 pKa = 4.33VQICWTNAGQGADD85 pKa = 3.5EE86 pKa = 4.32YY87 pKa = 11.23GKK89 pKa = 10.87FSTALEE95 pKa = 4.54AGTGAPDD102 pKa = 4.0VIQLEE107 pKa = 4.58SEE109 pKa = 4.0ILPTYY114 pKa = 9.67TILDD118 pKa = 4.17GLTDD122 pKa = 4.57LAPYY126 pKa = 9.98GADD129 pKa = 3.07EE130 pKa = 6.24LEE132 pKa = 4.31DD133 pKa = 3.73QFPAGAWSDD142 pKa = 3.81VSSGDD147 pKa = 3.48AVHH150 pKa = 7.54AIPVDD155 pKa = 3.94GGPVGMLYY163 pKa = 10.37RR164 pKa = 11.84ADD166 pKa = 3.33IFEE169 pKa = 4.74KK170 pKa = 10.95YY171 pKa = 9.94GVEE174 pKa = 5.28PPTTWDD180 pKa = 2.98EE181 pKa = 4.27FARR184 pKa = 11.84AAQDD188 pKa = 4.06LRR190 pKa = 11.84DD191 pKa = 3.79AGYY194 pKa = 10.85DD195 pKa = 3.72GYY197 pKa = 9.45ITNYY201 pKa = 7.9ATNGGAFNYY210 pKa = 10.85ALFAQAGWAPFDD222 pKa = 4.0YY223 pKa = 10.97DD224 pKa = 3.42PAEE227 pKa = 4.5RR228 pKa = 11.84DD229 pKa = 3.32QIGIDD234 pKa = 3.15VDD236 pKa = 3.89TPEE239 pKa = 3.92ARR241 pKa = 11.84QVLSYY246 pKa = 10.59WEE248 pKa = 4.24DD249 pKa = 4.09LIDD252 pKa = 4.55RR253 pKa = 11.84GLVSSDD259 pKa = 3.62DD260 pKa = 4.25AFTSDD265 pKa = 4.0YY266 pKa = 9.53NTSLVDD272 pKa = 3.05GTYY275 pKa = 10.93AVYY278 pKa = 10.06IAAAWGPGYY287 pKa = 10.93LEE289 pKa = 4.69GLSDD293 pKa = 4.54ADD295 pKa = 5.52SGAAWKK301 pKa = 9.5AAPLPQWDD309 pKa = 3.78PAEE312 pKa = 4.16PVQVNQGGSALAVTSQAKK330 pKa = 10.38DD331 pKa = 3.26PEE333 pKa = 4.33AAATVAMEE341 pKa = 4.5LFDD344 pKa = 6.62DD345 pKa = 4.24EE346 pKa = 4.76EE347 pKa = 4.19TWKK350 pKa = 10.68IGVEE354 pKa = 4.08DD355 pKa = 4.36AGLFPLWNLALDD367 pKa = 4.45ADD369 pKa = 4.16WFTDD373 pKa = 2.78RR374 pKa = 11.84RR375 pKa = 11.84YY376 pKa = 10.32PFFGGQQVNRR386 pKa = 11.84DD387 pKa = 3.64VFLDD391 pKa = 3.87AAAQYY396 pKa = 10.53PGFTFSPFQVYY407 pKa = 10.13AYY409 pKa = 10.25DD410 pKa = 3.8RR411 pKa = 11.84QTMALYY417 pKa = 11.17DD418 pKa = 4.0MVEE421 pKa = 4.7GPATAADD428 pKa = 3.77ALATVQDD435 pKa = 3.65SLVRR439 pKa = 11.84YY440 pKa = 9.84AEE442 pKa = 3.97QQGFTVRR449 pKa = 4.02
MM1 pKa = 7.78RR2 pKa = 11.84RR3 pKa = 11.84GLRR6 pKa = 11.84RR7 pKa = 11.84AVAASAAGLAGLSLAACAPGGDD29 pKa = 4.32GAPADD34 pKa = 3.79NTSCTNEE41 pKa = 3.88IKK43 pKa = 11.11VPDD46 pKa = 3.93ATRR49 pKa = 11.84VTLWAWYY56 pKa = 8.02PAVEE60 pKa = 4.59QIVDD64 pKa = 3.73TFNAQHH70 pKa = 7.48DD71 pKa = 4.24DD72 pKa = 4.33VQICWTNAGQGADD85 pKa = 3.5EE86 pKa = 4.32YY87 pKa = 11.23GKK89 pKa = 10.87FSTALEE95 pKa = 4.54AGTGAPDD102 pKa = 4.0VIQLEE107 pKa = 4.58SEE109 pKa = 4.0ILPTYY114 pKa = 9.67TILDD118 pKa = 4.17GLTDD122 pKa = 4.57LAPYY126 pKa = 9.98GADD129 pKa = 3.07EE130 pKa = 6.24LEE132 pKa = 4.31DD133 pKa = 3.73QFPAGAWSDD142 pKa = 3.81VSSGDD147 pKa = 3.48AVHH150 pKa = 7.54AIPVDD155 pKa = 3.94GGPVGMLYY163 pKa = 10.37RR164 pKa = 11.84ADD166 pKa = 3.33IFEE169 pKa = 4.74KK170 pKa = 10.95YY171 pKa = 9.94GVEE174 pKa = 5.28PPTTWDD180 pKa = 2.98EE181 pKa = 4.27FARR184 pKa = 11.84AAQDD188 pKa = 4.06LRR190 pKa = 11.84DD191 pKa = 3.79AGYY194 pKa = 10.85DD195 pKa = 3.72GYY197 pKa = 9.45ITNYY201 pKa = 7.9ATNGGAFNYY210 pKa = 10.85ALFAQAGWAPFDD222 pKa = 4.0YY223 pKa = 10.97DD224 pKa = 3.42PAEE227 pKa = 4.5RR228 pKa = 11.84DD229 pKa = 3.32QIGIDD234 pKa = 3.15VDD236 pKa = 3.89TPEE239 pKa = 3.92ARR241 pKa = 11.84QVLSYY246 pKa = 10.59WEE248 pKa = 4.24DD249 pKa = 4.09LIDD252 pKa = 4.55RR253 pKa = 11.84GLVSSDD259 pKa = 3.62DD260 pKa = 4.25AFTSDD265 pKa = 4.0YY266 pKa = 9.53NTSLVDD272 pKa = 3.05GTYY275 pKa = 10.93AVYY278 pKa = 10.06IAAAWGPGYY287 pKa = 10.93LEE289 pKa = 4.69GLSDD293 pKa = 4.54ADD295 pKa = 5.52SGAAWKK301 pKa = 9.5AAPLPQWDD309 pKa = 3.78PAEE312 pKa = 4.16PVQVNQGGSALAVTSQAKK330 pKa = 10.38DD331 pKa = 3.26PEE333 pKa = 4.33AAATVAMEE341 pKa = 4.5LFDD344 pKa = 6.62DD345 pKa = 4.24EE346 pKa = 4.76EE347 pKa = 4.19TWKK350 pKa = 10.68IGVEE354 pKa = 4.08DD355 pKa = 4.36AGLFPLWNLALDD367 pKa = 4.45ADD369 pKa = 4.16WFTDD373 pKa = 2.78RR374 pKa = 11.84RR375 pKa = 11.84YY376 pKa = 10.32PFFGGQQVNRR386 pKa = 11.84DD387 pKa = 3.64VFLDD391 pKa = 3.87AAAQYY396 pKa = 10.53PGFTFSPFQVYY407 pKa = 10.13AYY409 pKa = 10.25DD410 pKa = 3.8RR411 pKa = 11.84QTMALYY417 pKa = 11.17DD418 pKa = 4.0MVEE421 pKa = 4.7GPATAADD428 pKa = 3.77ALATVQDD435 pKa = 3.65SLVRR439 pKa = 11.84YY440 pKa = 9.84AEE442 pKa = 3.97QQGFTVRR449 pKa = 4.02
Molecular weight: 48.36 kDa
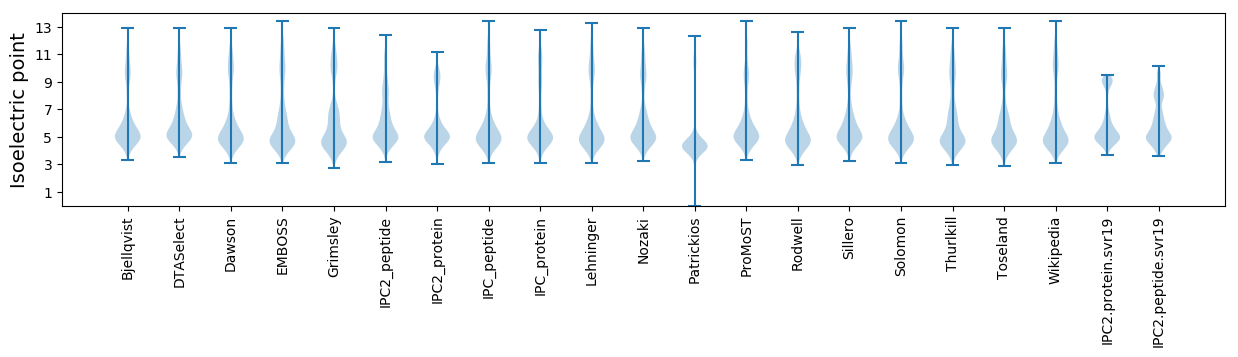
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1T5K9A5|A0A1T5K9A5_9MICO ArsR family transcriptional regulator OS=Krasilnikoviella flava OX=526729 GN=SAMN04324258_1958 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 10.24VRR4 pKa = 11.84ASLRR8 pKa = 11.84SLKK11 pKa = 10.12RR12 pKa = 11.84LPGAKK17 pKa = 8.89VVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84TFVINKK30 pKa = 7.51QNPRR34 pKa = 11.84FKK36 pKa = 10.8ARR38 pKa = 11.84QGG40 pKa = 3.28
MM1 pKa = 7.6KK2 pKa = 10.24VRR4 pKa = 11.84ASLRR8 pKa = 11.84SLKK11 pKa = 10.12RR12 pKa = 11.84LPGAKK17 pKa = 8.89VVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84TFVINKK30 pKa = 7.51QNPRR34 pKa = 11.84FKK36 pKa = 10.8ARR38 pKa = 11.84QGG40 pKa = 3.28
Molecular weight: 4.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1480017 |
37 |
2937 |
344.5 |
36.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.43 ± 0.063 | 0.506 ± 0.007 |
6.761 ± 0.035 | 5.325 ± 0.034 |
2.65 ± 0.023 | 9.63 ± 0.034 |
2.094 ± 0.02 | 2.705 ± 0.028 |
1.449 ± 0.023 | 10.208 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.604 ± 0.015 | 1.461 ± 0.018 |
6.017 ± 0.029 | 2.531 ± 0.02 |
7.834 ± 0.045 | 4.837 ± 0.025 |
6.322 ± 0.035 | 10.156 ± 0.041 |
1.604 ± 0.017 | 1.877 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
