
Paraburkholderia dinghuensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Paraburkholderia
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
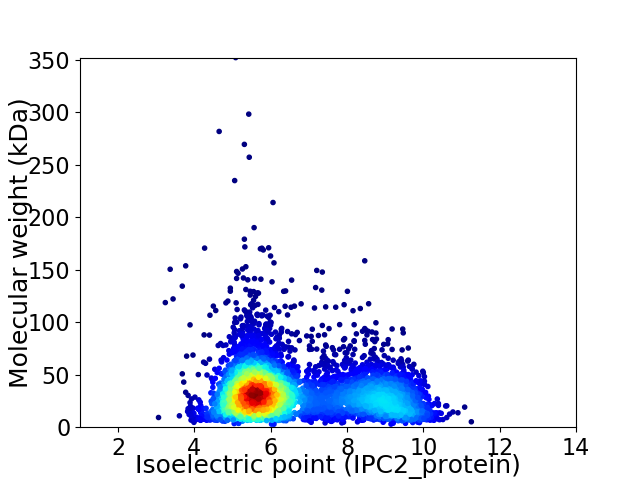
Virtual 2D-PAGE plot for 5170 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N6MN98|A0A3N6MN98_9BURK DUF1329 domain-containing protein OS=Paraburkholderia dinghuensis OX=2305225 GN=D1Y85_16685 PE=4 SV=1
LL1 pKa = 7.63DD2 pKa = 3.71NSAGGTLQTSSTDD15 pKa = 3.37LALASATIDD24 pKa = 3.47NDD26 pKa = 3.45GGTITHH32 pKa = 7.49AGTGTLTLVAGSGTGTLSNAGGTIASNGQVAAQAGAFDD70 pKa = 3.99NAAGAVIGQTGLTVTTRR87 pKa = 11.84NTLDD91 pKa = 3.34NTNGRR96 pKa = 11.84LLSNTDD102 pKa = 3.3LSLASSTLNNDD113 pKa = 3.37GGQIGAGANGAIQSGSLTNGGGSITASSLAVTTSGKK149 pKa = 10.22LDD151 pKa = 3.77NGSGDD156 pKa = 3.21IEE158 pKa = 4.42ANQLALNATDD168 pKa = 3.83LVNRR172 pKa = 11.84AGTITQYY179 pKa = 11.0GASPMSVNASGTFDD193 pKa = 3.41NSNGGTLQTNSTDD206 pKa = 3.1VTLAAGTIDD215 pKa = 3.84NDD217 pKa = 3.8GGTITHH223 pKa = 7.33AGAGTLTLDD232 pKa = 4.02ADD234 pKa = 4.06SGAGSLSNAGGTIASNGQVVARR256 pKa = 11.84SGAFGNEE263 pKa = 3.61AGTVIGQTGLSATVGNTLDD282 pKa = 3.66NTNGRR287 pKa = 11.84LLSNADD293 pKa = 3.47LSLASGTLDD302 pKa = 3.67NNGGQIGASANGTVQSGSLANSSGSIAASNLSVTAGTMLDD342 pKa = 3.46NSGGDD347 pKa = 3.17IEE349 pKa = 5.38ANQLALVATNLLNHH363 pKa = 7.07GGAITQYY370 pKa = 8.86GTSPMGFTVSGTFDD384 pKa = 3.27NSNGGTLQANSTDD397 pKa = 3.49LTLAAGTIDD406 pKa = 3.84NDD408 pKa = 3.8GGTITHH414 pKa = 7.49AGTGTLTLDD423 pKa = 3.7AGSGAGSLSNAGGTIASNGQAEE445 pKa = 4.33AQAGAFDD452 pKa = 4.03NAAGTVIGQTGLTATIGNTLDD473 pKa = 3.51NTNGRR478 pKa = 11.84LLSNADD484 pKa = 3.43LSLATGTLTNNGGQIGAAANGTIHH508 pKa = 6.81SGSLTNSGGSMVASNLSVTTATTLDD533 pKa = 3.58NSVGDD538 pKa = 3.72IEE540 pKa = 5.23ANQLALGATDD550 pKa = 4.77LLNHH554 pKa = 7.17GGTITQYY561 pKa = 10.57GASSMGFAVSGTFDD575 pKa = 3.34NSNGGMLQTNSTNLTLAPARR595 pKa = 11.84LDD597 pKa = 3.53NDD599 pKa = 3.43GGAILDD605 pKa = 4.4AGTGTLTIAPGNGAGSISNAGGQIITAGQIAAQAGSLDD643 pKa = 3.65NAGGVLAAQRR653 pKa = 11.84NIAANIAGNVNNTQGAIRR671 pKa = 11.84ALEE674 pKa = 4.11SLSLASGGALTNTNGQIQSGTGAGVGVSDD703 pKa = 5.17ASTLAIQAASIDD715 pKa = 3.86NTGGLVSNLGTGDD728 pKa = 3.36EE729 pKa = 4.53TVQGGSQTVNSGGVITGNGNVAVSTSSLSNTQGGQVSGANVAIQAGTVDD778 pKa = 3.62NSGGAIGTFAGPGGDD793 pKa = 3.43VGITASGTVTNTGGQIGATHH813 pKa = 7.1DD814 pKa = 3.92LSVNAAALTGGGTYY828 pKa = 10.99GAMNDD833 pKa = 3.45VAVTVQGNFTPTPDD847 pKa = 3.47LQFSAGHH854 pKa = 6.75DD855 pKa = 3.58LAFTLPGAFTNSATLDD871 pKa = 3.4AANNLDD877 pKa = 3.64VNANDD882 pKa = 4.23IEE884 pKa = 4.42NSGAMMAGGTLTTHH898 pKa = 6.94SATLEE903 pKa = 4.1NTGVMVGGSVSLNATQTLSNVGPTALIGATDD934 pKa = 3.78SNGLLEE940 pKa = 5.42LLAPDD945 pKa = 4.14IEE947 pKa = 4.48NRR949 pKa = 11.84DD950 pKa = 4.0DD951 pKa = 3.5TTMTDD956 pKa = 3.4TQATTAIYY964 pKa = 10.49GLGAMVLAGGKK975 pKa = 9.12DD976 pKa = 3.32ANGNYY981 pKa = 9.24TNANLIRR988 pKa = 11.84NQSGLIQSAGDD999 pKa = 3.32MTLAANQVTNTRR1011 pKa = 11.84TTMTTTGLNQPVDD1024 pKa = 4.13PDD1026 pKa = 4.25LLTQLGISLSGCTAVQVSDD1045 pKa = 4.7CDD1047 pKa = 3.64PGHH1050 pKa = 7.3PYY1052 pKa = 8.42VQRR1055 pKa = 11.84YY1056 pKa = 7.88QPDD1059 pKa = 3.46NPVFLTMIGGAFTMPPRR1076 pKa = 11.84SGQWNSNYY1084 pKa = 10.11QNTTYY1089 pKa = 10.54TGVALANLIASISPQAQIIAGGNLDD1114 pKa = 3.49ASKK1117 pKa = 11.16VGLFQNYY1124 pKa = 5.82WSAVAAVGNIALPVTLDD1141 pKa = 3.17QDD1143 pKa = 3.32SWQGQTAPGVQVTYY1157 pKa = 10.49SGYY1160 pKa = 9.4YY1161 pKa = 9.75HH1162 pKa = 6.6YY1163 pKa = 11.55ANYY1166 pKa = 9.87DD1167 pKa = 3.07QSIRR1171 pKa = 11.84DD1172 pKa = 3.39WTLPFGDD1179 pKa = 4.25APFVGSNPGGYY1190 pKa = 7.19TQAAPADD1197 pKa = 3.77VRR1199 pKa = 11.84TYY1201 pKa = 11.33ALPSYY1206 pKa = 10.18EE1207 pKa = 4.22STFVAGGTISGTGVSINNTAGNAGVPSLGLVPGQSVGGVNAGGVSGSADD1256 pKa = 2.95GSAAGGTSAIQGSAGGSNTGATSVVQGSAGSPDD1289 pKa = 3.17SAAASVSGSTGLTNPVIASATAVNVLSNLTIPQGGLFRR1327 pKa = 11.84PATAPNAQYY1336 pKa = 10.93LIEE1339 pKa = 4.42TNPAFTSQKK1348 pKa = 10.72SFISSDD1354 pKa = 3.75YY1355 pKa = 10.82YY1356 pKa = 11.06LQQLGLNPQTTEE1368 pKa = 3.48KK1369 pKa = 10.88RR1370 pKa = 11.84LGDD1373 pKa = 3.66GLYY1376 pKa = 9.89EE1377 pKa = 4.06QQLVV1381 pKa = 3.4
LL1 pKa = 7.63DD2 pKa = 3.71NSAGGTLQTSSTDD15 pKa = 3.37LALASATIDD24 pKa = 3.47NDD26 pKa = 3.45GGTITHH32 pKa = 7.49AGTGTLTLVAGSGTGTLSNAGGTIASNGQVAAQAGAFDD70 pKa = 3.99NAAGAVIGQTGLTVTTRR87 pKa = 11.84NTLDD91 pKa = 3.34NTNGRR96 pKa = 11.84LLSNTDD102 pKa = 3.3LSLASSTLNNDD113 pKa = 3.37GGQIGAGANGAIQSGSLTNGGGSITASSLAVTTSGKK149 pKa = 10.22LDD151 pKa = 3.77NGSGDD156 pKa = 3.21IEE158 pKa = 4.42ANQLALNATDD168 pKa = 3.83LVNRR172 pKa = 11.84AGTITQYY179 pKa = 11.0GASPMSVNASGTFDD193 pKa = 3.41NSNGGTLQTNSTDD206 pKa = 3.1VTLAAGTIDD215 pKa = 3.84NDD217 pKa = 3.8GGTITHH223 pKa = 7.33AGAGTLTLDD232 pKa = 4.02ADD234 pKa = 4.06SGAGSLSNAGGTIASNGQVVARR256 pKa = 11.84SGAFGNEE263 pKa = 3.61AGTVIGQTGLSATVGNTLDD282 pKa = 3.66NTNGRR287 pKa = 11.84LLSNADD293 pKa = 3.47LSLASGTLDD302 pKa = 3.67NNGGQIGASANGTVQSGSLANSSGSIAASNLSVTAGTMLDD342 pKa = 3.46NSGGDD347 pKa = 3.17IEE349 pKa = 5.38ANQLALVATNLLNHH363 pKa = 7.07GGAITQYY370 pKa = 8.86GTSPMGFTVSGTFDD384 pKa = 3.27NSNGGTLQANSTDD397 pKa = 3.49LTLAAGTIDD406 pKa = 3.84NDD408 pKa = 3.8GGTITHH414 pKa = 7.49AGTGTLTLDD423 pKa = 3.7AGSGAGSLSNAGGTIASNGQAEE445 pKa = 4.33AQAGAFDD452 pKa = 4.03NAAGTVIGQTGLTATIGNTLDD473 pKa = 3.51NTNGRR478 pKa = 11.84LLSNADD484 pKa = 3.43LSLATGTLTNNGGQIGAAANGTIHH508 pKa = 6.81SGSLTNSGGSMVASNLSVTTATTLDD533 pKa = 3.58NSVGDD538 pKa = 3.72IEE540 pKa = 5.23ANQLALGATDD550 pKa = 4.77LLNHH554 pKa = 7.17GGTITQYY561 pKa = 10.57GASSMGFAVSGTFDD575 pKa = 3.34NSNGGMLQTNSTNLTLAPARR595 pKa = 11.84LDD597 pKa = 3.53NDD599 pKa = 3.43GGAILDD605 pKa = 4.4AGTGTLTIAPGNGAGSISNAGGQIITAGQIAAQAGSLDD643 pKa = 3.65NAGGVLAAQRR653 pKa = 11.84NIAANIAGNVNNTQGAIRR671 pKa = 11.84ALEE674 pKa = 4.11SLSLASGGALTNTNGQIQSGTGAGVGVSDD703 pKa = 5.17ASTLAIQAASIDD715 pKa = 3.86NTGGLVSNLGTGDD728 pKa = 3.36EE729 pKa = 4.53TVQGGSQTVNSGGVITGNGNVAVSTSSLSNTQGGQVSGANVAIQAGTVDD778 pKa = 3.62NSGGAIGTFAGPGGDD793 pKa = 3.43VGITASGTVTNTGGQIGATHH813 pKa = 7.1DD814 pKa = 3.92LSVNAAALTGGGTYY828 pKa = 10.99GAMNDD833 pKa = 3.45VAVTVQGNFTPTPDD847 pKa = 3.47LQFSAGHH854 pKa = 6.75DD855 pKa = 3.58LAFTLPGAFTNSATLDD871 pKa = 3.4AANNLDD877 pKa = 3.64VNANDD882 pKa = 4.23IEE884 pKa = 4.42NSGAMMAGGTLTTHH898 pKa = 6.94SATLEE903 pKa = 4.1NTGVMVGGSVSLNATQTLSNVGPTALIGATDD934 pKa = 3.78SNGLLEE940 pKa = 5.42LLAPDD945 pKa = 4.14IEE947 pKa = 4.48NRR949 pKa = 11.84DD950 pKa = 4.0DD951 pKa = 3.5TTMTDD956 pKa = 3.4TQATTAIYY964 pKa = 10.49GLGAMVLAGGKK975 pKa = 9.12DD976 pKa = 3.32ANGNYY981 pKa = 9.24TNANLIRR988 pKa = 11.84NQSGLIQSAGDD999 pKa = 3.32MTLAANQVTNTRR1011 pKa = 11.84TTMTTTGLNQPVDD1024 pKa = 4.13PDD1026 pKa = 4.25LLTQLGISLSGCTAVQVSDD1045 pKa = 4.7CDD1047 pKa = 3.64PGHH1050 pKa = 7.3PYY1052 pKa = 8.42VQRR1055 pKa = 11.84YY1056 pKa = 7.88QPDD1059 pKa = 3.46NPVFLTMIGGAFTMPPRR1076 pKa = 11.84SGQWNSNYY1084 pKa = 10.11QNTTYY1089 pKa = 10.54TGVALANLIASISPQAQIIAGGNLDD1114 pKa = 3.49ASKK1117 pKa = 11.16VGLFQNYY1124 pKa = 5.82WSAVAAVGNIALPVTLDD1141 pKa = 3.17QDD1143 pKa = 3.32SWQGQTAPGVQVTYY1157 pKa = 10.49SGYY1160 pKa = 9.4YY1161 pKa = 9.75HH1162 pKa = 6.6YY1163 pKa = 11.55ANYY1166 pKa = 9.87DD1167 pKa = 3.07QSIRR1171 pKa = 11.84DD1172 pKa = 3.39WTLPFGDD1179 pKa = 4.25APFVGSNPGGYY1190 pKa = 7.19TQAAPADD1197 pKa = 3.77VRR1199 pKa = 11.84TYY1201 pKa = 11.33ALPSYY1206 pKa = 10.18EE1207 pKa = 4.22STFVAGGTISGTGVSINNTAGNAGVPSLGLVPGQSVGGVNAGGVSGSADD1256 pKa = 2.95GSAAGGTSAIQGSAGGSNTGATSVVQGSAGSPDD1289 pKa = 3.17SAAASVSGSTGLTNPVIASATAVNVLSNLTIPQGGLFRR1327 pKa = 11.84PATAPNAQYY1336 pKa = 10.93LIEE1339 pKa = 4.42TNPAFTSQKK1348 pKa = 10.72SFISSDD1354 pKa = 3.75YY1355 pKa = 10.82YY1356 pKa = 11.06LQQLGLNPQTTEE1368 pKa = 3.48KK1369 pKa = 10.88RR1370 pKa = 11.84LGDD1373 pKa = 3.66GLYY1376 pKa = 9.89EE1377 pKa = 4.06QQLVV1381 pKa = 3.4
Molecular weight: 134.42 kDa
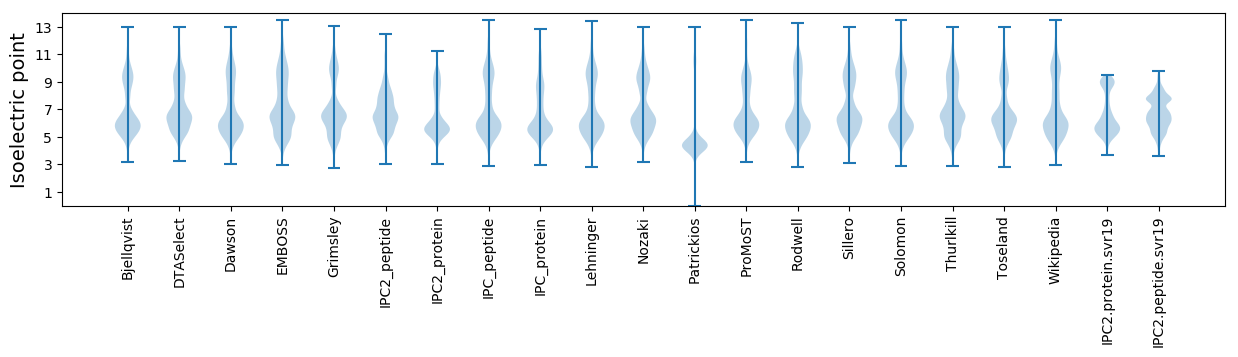
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N6N6N1|A0A3N6N6N1_9BURK Rod shape-determining protein MreD OS=Paraburkholderia dinghuensis OX=2305225 GN=mreD PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.74TAGGRR28 pKa = 11.84KK29 pKa = 9.04VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.74TAGGRR28 pKa = 11.84KK29 pKa = 9.04VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1685335 |
26 |
3298 |
326.0 |
35.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.585 ± 0.048 | 0.957 ± 0.01 |
5.457 ± 0.027 | 5.171 ± 0.032 |
3.682 ± 0.021 | 8.205 ± 0.035 |
2.329 ± 0.015 | 4.711 ± 0.021 |
3.057 ± 0.032 | 10.033 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.356 ± 0.016 | 2.901 ± 0.024 |
5.09 ± 0.029 | 3.638 ± 0.024 |
6.9 ± 0.04 | 5.71 ± 0.051 |
5.55 ± 0.033 | 7.833 ± 0.027 |
1.394 ± 0.016 | 2.441 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
