
Marinoscillum furvescens DSM 4134
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Reichenbachiellaceae; Marinoscillum; Marinoscillum furvescens
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
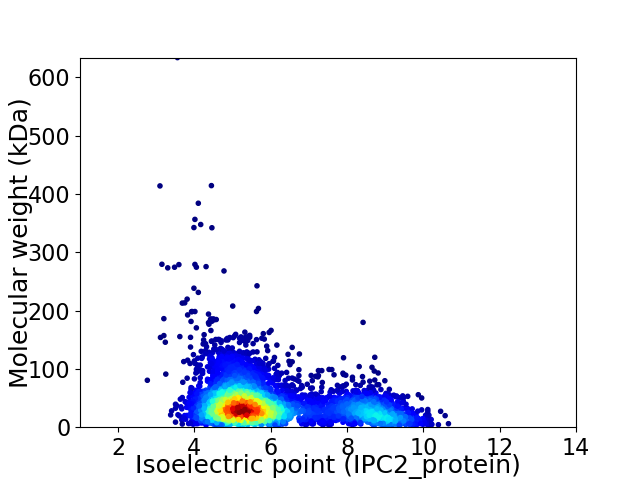
Virtual 2D-PAGE plot for 4702 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3D9L985|A0A3D9L985_9BACT Uncharacterized protein OS=Marinoscillum furvescens DSM 4134 OX=1122208 GN=C7460_103139 PE=4 SV=1
MM1 pKa = 7.41NNMKK5 pKa = 10.31YY6 pKa = 9.93IYY8 pKa = 10.11SALFLGLFSIALAQKK23 pKa = 9.7PIVVSLDD30 pKa = 3.28KK31 pKa = 11.46YY32 pKa = 10.68SATVDD37 pKa = 3.08EE38 pKa = 5.19TIFINGANFPTNVANVSVFFGNGKK62 pKa = 8.8ATVTSAAEE70 pKa = 3.87NQLQVTVTNTATFGPVRR87 pKa = 11.84VVNTSNGLAGASGQPFMLSYY107 pKa = 11.06SGDD110 pKa = 3.56ALDD113 pKa = 3.91ASKK116 pKa = 10.3FVKK119 pKa = 10.45RR120 pKa = 11.84STTTSEE126 pKa = 3.34QGTYY130 pKa = 10.06DD131 pKa = 3.59VCVCDD136 pKa = 5.59FNDD139 pKa = 4.3NGKK142 pKa = 10.02LDD144 pKa = 3.68AAVANFNNRR153 pKa = 11.84NISIFNNLTTFGNQASFSKK172 pKa = 10.83SSLDD176 pKa = 3.49NVSPTISVEE185 pKa = 4.05CGDD188 pKa = 4.52LNGDD192 pKa = 4.04GLPDD196 pKa = 3.78LVATSFSGSAPQHH209 pKa = 5.26VKK211 pKa = 10.34VYY213 pKa = 10.46QNNGTAGISFTKK225 pKa = 10.38VLEE228 pKa = 4.02FRR230 pKa = 11.84LPNLSASVPRR240 pKa = 11.84NPRR243 pKa = 11.84KK244 pKa = 9.81IKK246 pKa = 10.31IADD249 pKa = 3.4MDD251 pKa = 4.63LDD253 pKa = 4.23GKK255 pKa = 10.98SDD257 pKa = 4.76LIIGSEE263 pKa = 3.97NDD265 pKa = 3.06NSIFIYY271 pKa = 10.81QNTTTGGTLSFASSPEE287 pKa = 4.17SITVSGADD295 pKa = 3.07NAASIEE301 pKa = 4.08IGDD304 pKa = 4.53LNNDD308 pKa = 3.9GLPDD312 pKa = 3.62IAAVALNIPNSSVFVIRR329 pKa = 11.84NQSSPGSFNFIQASTINDD347 pKa = 3.34SNQRR351 pKa = 11.84TNVSLVDD358 pKa = 3.21IDD360 pKa = 5.34GDD362 pKa = 4.06FKK364 pKa = 11.58PEE366 pKa = 4.19IITTNNLKK374 pKa = 10.45NDD376 pKa = 2.82VDD378 pKa = 3.5IFEE381 pKa = 4.67NTTSGIEE388 pKa = 3.74ITFDD392 pKa = 3.37NSPTTLSVQRR402 pKa = 11.84PWSVASGDD410 pKa = 3.79LNGDD414 pKa = 3.58GKK416 pKa = 11.39VDD418 pKa = 4.01LSVATLSADD427 pKa = 2.95IYY429 pKa = 10.64VLEE432 pKa = 4.4NTTSNGSIAFTSSTFSATPDD452 pKa = 3.12NRR454 pKa = 11.84NIEE457 pKa = 4.06LADD460 pKa = 3.36IDD462 pKa = 4.99GDD464 pKa = 3.96AKK466 pKa = 10.86PDD468 pKa = 3.73VITTNNTGEE477 pKa = 4.27TATGSLLVMLNQNCTVSKK495 pKa = 10.49ILPEE499 pKa = 4.59DD500 pKa = 3.21LTFCLNEE507 pKa = 4.87PITLRR512 pKa = 11.84ATNTRR517 pKa = 11.84NATYY521 pKa = 10.16SWAITSGNGNITGSGQEE538 pKa = 3.89VQVTVTSGEE547 pKa = 4.21TATVQVTTAANDD559 pKa = 4.39GSCSEE564 pKa = 4.63ASSQVFSLTGGTPPPTPSISNSASGTICSGDD595 pKa = 3.76SFTLTASTATADD607 pKa = 3.35EE608 pKa = 5.13YY609 pKa = 11.79YY610 pKa = 9.83WVTPSGDD617 pKa = 3.62QITTTTNTLEE627 pKa = 4.19FSDD630 pKa = 4.74VSSADD635 pKa = 3.01AGNYY639 pKa = 6.2TVRR642 pKa = 11.84TFTSGSCTSAEE653 pKa = 4.25SPALTVAIDD662 pKa = 3.79EE663 pKa = 4.78PPSVVIASSASTTFCANSTINLIVPDD689 pKa = 4.1YY690 pKa = 11.21DD691 pKa = 3.57GYY693 pKa = 11.22SYY695 pKa = 10.9QWHH698 pKa = 7.25KK699 pKa = 11.42DD700 pKa = 3.29GTDD703 pKa = 3.54LTGEE707 pKa = 4.1TANTFTANASGSYY720 pKa = 9.12TVTLTSNATSCSKK733 pKa = 10.02TSGAIALTAVEE744 pKa = 4.73LPVGNIASEE753 pKa = 4.88DD754 pKa = 4.09EE755 pKa = 4.09ICVNAEE761 pKa = 3.42ISFDD765 pKa = 3.82GSGSTSDD772 pKa = 3.69PNGEE776 pKa = 4.24LTYY779 pKa = 11.0SWDD782 pKa = 3.73FGDD785 pKa = 5.41GNTADD790 pKa = 4.13TEE792 pKa = 4.79TANHH796 pKa = 7.08TYY798 pKa = 7.88TTDD801 pKa = 2.9GSYY804 pKa = 9.46TVTLTTGYY812 pKa = 11.01SNVNSCEE819 pKa = 3.87STTQKK824 pKa = 9.43TVEE827 pKa = 4.02IAALPSNDD835 pKa = 3.98EE836 pKa = 3.74IAAALTPDD844 pKa = 3.62PSTTQKK850 pKa = 10.72CPEE853 pKa = 4.54DD854 pKa = 4.1SLTLSLPTNYY864 pKa = 9.87LAYY867 pKa = 9.65EE868 pKa = 4.03WTVDD872 pKa = 3.19GDD874 pKa = 4.31VVSTTNSAKK883 pKa = 10.38IGTAPKK889 pKa = 10.39SSSTDD894 pKa = 2.98VTIDD898 pKa = 3.07VTTDD902 pKa = 3.48LGCVVDD908 pKa = 3.77ATVITVSNLANAGFTFSSPDD928 pKa = 3.32AQIVNDD934 pKa = 5.2TITLDD939 pKa = 3.5DD940 pKa = 3.62RR941 pKa = 11.84TAEE944 pKa = 4.06VTLQVSNGTDD954 pKa = 3.96FTWSPEE960 pKa = 3.92EE961 pKa = 4.41VISSTSGEE969 pKa = 4.35MIDD972 pKa = 3.59VYY974 pKa = 10.82PRR976 pKa = 11.84SQYY979 pKa = 8.05TTVTVTGTDD988 pKa = 3.38TEE990 pKa = 4.94GCTNTSQITIVSPGLIPRR1008 pKa = 11.84KK1009 pKa = 9.8SFSPNGDD1016 pKa = 3.58GIGYY1020 pKa = 7.54EE1021 pKa = 4.06CWEE1024 pKa = 4.17ILNSEE1029 pKa = 4.53SLTGCTIFILDD1040 pKa = 3.46QKK1042 pKa = 10.72GSHH1045 pKa = 5.19VLKK1048 pKa = 10.98AEE1050 pKa = 3.92APFVDD1055 pKa = 3.24NCVWNGNIDD1064 pKa = 3.69NGTSPVPEE1072 pKa = 3.76GVYY1075 pKa = 10.53YY1076 pKa = 10.62YY1077 pKa = 10.11ILKK1080 pKa = 10.38CDD1082 pKa = 4.47DD1083 pKa = 4.37GADD1086 pKa = 3.79SSTGTIMLARR1096 pKa = 4.46
MM1 pKa = 7.41NNMKK5 pKa = 10.31YY6 pKa = 9.93IYY8 pKa = 10.11SALFLGLFSIALAQKK23 pKa = 9.7PIVVSLDD30 pKa = 3.28KK31 pKa = 11.46YY32 pKa = 10.68SATVDD37 pKa = 3.08EE38 pKa = 5.19TIFINGANFPTNVANVSVFFGNGKK62 pKa = 8.8ATVTSAAEE70 pKa = 3.87NQLQVTVTNTATFGPVRR87 pKa = 11.84VVNTSNGLAGASGQPFMLSYY107 pKa = 11.06SGDD110 pKa = 3.56ALDD113 pKa = 3.91ASKK116 pKa = 10.3FVKK119 pKa = 10.45RR120 pKa = 11.84STTTSEE126 pKa = 3.34QGTYY130 pKa = 10.06DD131 pKa = 3.59VCVCDD136 pKa = 5.59FNDD139 pKa = 4.3NGKK142 pKa = 10.02LDD144 pKa = 3.68AAVANFNNRR153 pKa = 11.84NISIFNNLTTFGNQASFSKK172 pKa = 10.83SSLDD176 pKa = 3.49NVSPTISVEE185 pKa = 4.05CGDD188 pKa = 4.52LNGDD192 pKa = 4.04GLPDD196 pKa = 3.78LVATSFSGSAPQHH209 pKa = 5.26VKK211 pKa = 10.34VYY213 pKa = 10.46QNNGTAGISFTKK225 pKa = 10.38VLEE228 pKa = 4.02FRR230 pKa = 11.84LPNLSASVPRR240 pKa = 11.84NPRR243 pKa = 11.84KK244 pKa = 9.81IKK246 pKa = 10.31IADD249 pKa = 3.4MDD251 pKa = 4.63LDD253 pKa = 4.23GKK255 pKa = 10.98SDD257 pKa = 4.76LIIGSEE263 pKa = 3.97NDD265 pKa = 3.06NSIFIYY271 pKa = 10.81QNTTTGGTLSFASSPEE287 pKa = 4.17SITVSGADD295 pKa = 3.07NAASIEE301 pKa = 4.08IGDD304 pKa = 4.53LNNDD308 pKa = 3.9GLPDD312 pKa = 3.62IAAVALNIPNSSVFVIRR329 pKa = 11.84NQSSPGSFNFIQASTINDD347 pKa = 3.34SNQRR351 pKa = 11.84TNVSLVDD358 pKa = 3.21IDD360 pKa = 5.34GDD362 pKa = 4.06FKK364 pKa = 11.58PEE366 pKa = 4.19IITTNNLKK374 pKa = 10.45NDD376 pKa = 2.82VDD378 pKa = 3.5IFEE381 pKa = 4.67NTTSGIEE388 pKa = 3.74ITFDD392 pKa = 3.37NSPTTLSVQRR402 pKa = 11.84PWSVASGDD410 pKa = 3.79LNGDD414 pKa = 3.58GKK416 pKa = 11.39VDD418 pKa = 4.01LSVATLSADD427 pKa = 2.95IYY429 pKa = 10.64VLEE432 pKa = 4.4NTTSNGSIAFTSSTFSATPDD452 pKa = 3.12NRR454 pKa = 11.84NIEE457 pKa = 4.06LADD460 pKa = 3.36IDD462 pKa = 4.99GDD464 pKa = 3.96AKK466 pKa = 10.86PDD468 pKa = 3.73VITTNNTGEE477 pKa = 4.27TATGSLLVMLNQNCTVSKK495 pKa = 10.49ILPEE499 pKa = 4.59DD500 pKa = 3.21LTFCLNEE507 pKa = 4.87PITLRR512 pKa = 11.84ATNTRR517 pKa = 11.84NATYY521 pKa = 10.16SWAITSGNGNITGSGQEE538 pKa = 3.89VQVTVTSGEE547 pKa = 4.21TATVQVTTAANDD559 pKa = 4.39GSCSEE564 pKa = 4.63ASSQVFSLTGGTPPPTPSISNSASGTICSGDD595 pKa = 3.76SFTLTASTATADD607 pKa = 3.35EE608 pKa = 5.13YY609 pKa = 11.79YY610 pKa = 9.83WVTPSGDD617 pKa = 3.62QITTTTNTLEE627 pKa = 4.19FSDD630 pKa = 4.74VSSADD635 pKa = 3.01AGNYY639 pKa = 6.2TVRR642 pKa = 11.84TFTSGSCTSAEE653 pKa = 4.25SPALTVAIDD662 pKa = 3.79EE663 pKa = 4.78PPSVVIASSASTTFCANSTINLIVPDD689 pKa = 4.1YY690 pKa = 11.21DD691 pKa = 3.57GYY693 pKa = 11.22SYY695 pKa = 10.9QWHH698 pKa = 7.25KK699 pKa = 11.42DD700 pKa = 3.29GTDD703 pKa = 3.54LTGEE707 pKa = 4.1TANTFTANASGSYY720 pKa = 9.12TVTLTSNATSCSKK733 pKa = 10.02TSGAIALTAVEE744 pKa = 4.73LPVGNIASEE753 pKa = 4.88DD754 pKa = 4.09EE755 pKa = 4.09ICVNAEE761 pKa = 3.42ISFDD765 pKa = 3.82GSGSTSDD772 pKa = 3.69PNGEE776 pKa = 4.24LTYY779 pKa = 11.0SWDD782 pKa = 3.73FGDD785 pKa = 5.41GNTADD790 pKa = 4.13TEE792 pKa = 4.79TANHH796 pKa = 7.08TYY798 pKa = 7.88TTDD801 pKa = 2.9GSYY804 pKa = 9.46TVTLTTGYY812 pKa = 11.01SNVNSCEE819 pKa = 3.87STTQKK824 pKa = 9.43TVEE827 pKa = 4.02IAALPSNDD835 pKa = 3.98EE836 pKa = 3.74IAAALTPDD844 pKa = 3.62PSTTQKK850 pKa = 10.72CPEE853 pKa = 4.54DD854 pKa = 4.1SLTLSLPTNYY864 pKa = 9.87LAYY867 pKa = 9.65EE868 pKa = 4.03WTVDD872 pKa = 3.19GDD874 pKa = 4.31VVSTTNSAKK883 pKa = 10.38IGTAPKK889 pKa = 10.39SSSTDD894 pKa = 2.98VTIDD898 pKa = 3.07VTTDD902 pKa = 3.48LGCVVDD908 pKa = 3.77ATVITVSNLANAGFTFSSPDD928 pKa = 3.32AQIVNDD934 pKa = 5.2TITLDD939 pKa = 3.5DD940 pKa = 3.62RR941 pKa = 11.84TAEE944 pKa = 4.06VTLQVSNGTDD954 pKa = 3.96FTWSPEE960 pKa = 3.92EE961 pKa = 4.41VISSTSGEE969 pKa = 4.35MIDD972 pKa = 3.59VYY974 pKa = 10.82PRR976 pKa = 11.84SQYY979 pKa = 8.05TTVTVTGTDD988 pKa = 3.38TEE990 pKa = 4.94GCTNTSQITIVSPGLIPRR1008 pKa = 11.84KK1009 pKa = 9.8SFSPNGDD1016 pKa = 3.58GIGYY1020 pKa = 7.54EE1021 pKa = 4.06CWEE1024 pKa = 4.17ILNSEE1029 pKa = 4.53SLTGCTIFILDD1040 pKa = 3.46QKK1042 pKa = 10.72GSHH1045 pKa = 5.19VLKK1048 pKa = 10.98AEE1050 pKa = 3.92APFVDD1055 pKa = 3.24NCVWNGNIDD1064 pKa = 3.69NGTSPVPEE1072 pKa = 3.76GVYY1075 pKa = 10.53YY1076 pKa = 10.62YY1077 pKa = 10.11ILKK1080 pKa = 10.38CDD1082 pKa = 4.47DD1083 pKa = 4.37GADD1086 pKa = 3.79SSTGTIMLARR1096 pKa = 4.46
Molecular weight: 114.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3D9L4V2|A0A3D9L4V2_9BACT Uridylate kinase OS=Marinoscillum furvescens DSM 4134 OX=1122208 GN=pyrH PE=3 SV=1
MM1 pKa = 7.82PLFYY5 pKa = 10.22WCALVFKK12 pKa = 10.4LRR14 pKa = 11.84TGLEE18 pKa = 4.0DD19 pKa = 3.36TFVKK23 pKa = 10.26IFPVAAIFRR32 pKa = 11.84VNAATVRR39 pKa = 11.84PNAGVHH45 pKa = 5.85PMNAAIPHH53 pKa = 6.17INAEE57 pKa = 4.25TFRR60 pKa = 11.84ISAIIVRR67 pKa = 11.84VNAEE71 pKa = 4.22TLRR74 pKa = 11.84MNAAIVRR81 pKa = 11.84PNTVVHH87 pKa = 6.42PMNVAIPHH95 pKa = 6.26INAEE99 pKa = 4.37TFRR102 pKa = 11.84MNAIAVHH109 pKa = 5.93VNAEE113 pKa = 4.3TLRR116 pKa = 11.84MNAAIFRR123 pKa = 11.84ISSTIGLSEE132 pKa = 3.93
MM1 pKa = 7.82PLFYY5 pKa = 10.22WCALVFKK12 pKa = 10.4LRR14 pKa = 11.84TGLEE18 pKa = 4.0DD19 pKa = 3.36TFVKK23 pKa = 10.26IFPVAAIFRR32 pKa = 11.84VNAATVRR39 pKa = 11.84PNAGVHH45 pKa = 5.85PMNAAIPHH53 pKa = 6.17INAEE57 pKa = 4.25TFRR60 pKa = 11.84ISAIIVRR67 pKa = 11.84VNAEE71 pKa = 4.22TLRR74 pKa = 11.84MNAAIVRR81 pKa = 11.84PNTVVHH87 pKa = 6.42PMNVAIPHH95 pKa = 6.26INAEE99 pKa = 4.37TFRR102 pKa = 11.84MNAIAVHH109 pKa = 5.93VNAEE113 pKa = 4.3TLRR116 pKa = 11.84MNAAIFRR123 pKa = 11.84ISSTIGLSEE132 pKa = 3.93
Molecular weight: 14.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1798820 |
29 |
6013 |
382.6 |
42.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.379 ± 0.035 | 0.724 ± 0.01 |
5.982 ± 0.043 | 6.613 ± 0.034 |
4.667 ± 0.026 | 7.188 ± 0.041 |
2.071 ± 0.019 | 6.363 ± 0.03 |
5.736 ± 0.042 | 9.413 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.227 ± 0.016 | 4.967 ± 0.033 |
3.892 ± 0.022 | 3.922 ± 0.022 |
4.132 ± 0.029 | 6.724 ± 0.037 |
5.872 ± 0.048 | 6.673 ± 0.028 |
1.358 ± 0.014 | 4.097 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
