
Hibiscus latent Fort Pierce virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
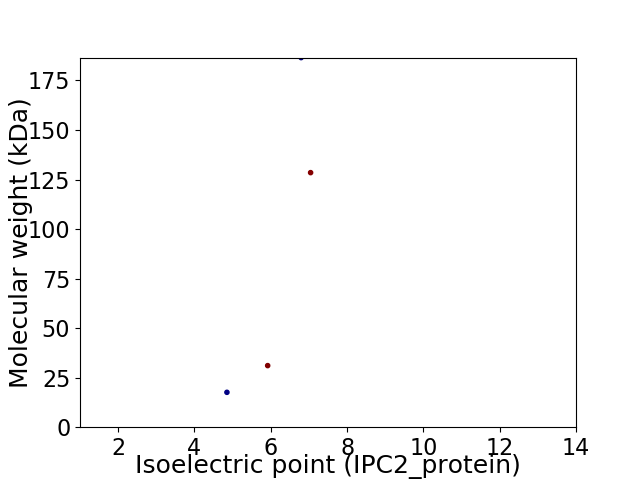
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A097ZNB1|A0A097ZNB1_9VIRU 31 kDa movement protein OS=Hibiscus latent Fort Pierce virus OX=233051 PE=4 SV=1
MM1 pKa = 7.57SYY3 pKa = 11.7SNITALNLIYY13 pKa = 10.45SSPTFAPYY21 pKa = 10.45DD22 pKa = 3.59VLLEE26 pKa = 3.86ILIKK30 pKa = 10.67SRR32 pKa = 11.84SNSFQTQAGRR42 pKa = 11.84DD43 pKa = 3.44ILRR46 pKa = 11.84EE47 pKa = 3.79QLVNALQIVVTPTTRR62 pKa = 11.84FPADD66 pKa = 2.77RR67 pKa = 11.84WYY69 pKa = 10.26VWSRR73 pKa = 11.84NPTLGPVFEE82 pKa = 5.67ALLQATDD89 pKa = 3.35TKK91 pKa = 11.33NRR93 pKa = 11.84IIEE96 pKa = 4.26TEE98 pKa = 3.84EE99 pKa = 3.89DD100 pKa = 3.67SRR102 pKa = 11.84PTTAEE107 pKa = 3.93TLNATQRR114 pKa = 11.84VDD116 pKa = 3.35DD117 pKa = 4.06ATVAIHH123 pKa = 7.12KK124 pKa = 9.93EE125 pKa = 3.3IDD127 pKa = 3.73NILGLLQGGTAVYY140 pKa = 9.81NQASFEE146 pKa = 4.19IASGWTWTAPANN158 pKa = 3.7
MM1 pKa = 7.57SYY3 pKa = 11.7SNITALNLIYY13 pKa = 10.45SSPTFAPYY21 pKa = 10.45DD22 pKa = 3.59VLLEE26 pKa = 3.86ILIKK30 pKa = 10.67SRR32 pKa = 11.84SNSFQTQAGRR42 pKa = 11.84DD43 pKa = 3.44ILRR46 pKa = 11.84EE47 pKa = 3.79QLVNALQIVVTPTTRR62 pKa = 11.84FPADD66 pKa = 2.77RR67 pKa = 11.84WYY69 pKa = 10.26VWSRR73 pKa = 11.84NPTLGPVFEE82 pKa = 5.67ALLQATDD89 pKa = 3.35TKK91 pKa = 11.33NRR93 pKa = 11.84IIEE96 pKa = 4.26TEE98 pKa = 3.84EE99 pKa = 3.89DD100 pKa = 3.67SRR102 pKa = 11.84PTTAEE107 pKa = 3.93TLNATQRR114 pKa = 11.84VDD116 pKa = 3.35DD117 pKa = 4.06ATVAIHH123 pKa = 7.12KK124 pKa = 9.93EE125 pKa = 3.3IDD127 pKa = 3.73NILGLLQGGTAVYY140 pKa = 9.81NQASFEE146 pKa = 4.19IASGWTWTAPANN158 pKa = 3.7
Molecular weight: 17.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B6E277|B6E277_9VIRU 128 kDa replicase OS=Hibiscus latent Fort Pierce virus OX=233051 PE=4 SV=1
MM1 pKa = 6.62TTSTTDD7 pKa = 2.67IKK9 pKa = 10.91QQITNVRR16 pKa = 11.84DD17 pKa = 3.73DD18 pKa = 4.12AAHH21 pKa = 6.6GAASLIKK28 pKa = 10.28GLATRR33 pKa = 11.84RR34 pKa = 11.84VYY36 pKa = 11.08DD37 pKa = 3.58EE38 pKa = 4.64AVNTLTLLDD47 pKa = 4.25KK48 pKa = 10.73RR49 pKa = 11.84PKK51 pKa = 10.21FSFSNLVSTEE61 pKa = 3.67NAKK64 pKa = 9.42MVTEE68 pKa = 5.5AYY70 pKa = 9.73PEE72 pKa = 4.04FNISFVGNRR81 pKa = 11.84NTVHH85 pKa = 6.58SLAGGLRR92 pKa = 11.84KK93 pKa = 9.93LEE95 pKa = 4.02MEE97 pKa = 4.48YY98 pKa = 11.29LMTLVPYY105 pKa = 10.68GSATFDD111 pKa = 2.71IGGNYY116 pKa = 8.98AQHH119 pKa = 6.51LLKK122 pKa = 10.75GRR124 pKa = 11.84AYY126 pKa = 8.8VHH128 pKa = 6.65CCNPCLDD135 pKa = 3.98FRR137 pKa = 11.84DD138 pKa = 3.57IARR141 pKa = 11.84NEE143 pKa = 4.27SYY145 pKa = 11.08KK146 pKa = 10.53DD147 pKa = 3.53TVDD150 pKa = 3.8SYY152 pKa = 11.66LKK154 pKa = 10.69RR155 pKa = 11.84FTTPDD160 pKa = 3.61CGWFSGAVRR169 pKa = 11.84AITARR174 pKa = 11.84PLPSFQKK181 pKa = 10.5EE182 pKa = 3.52AFEE185 pKa = 5.24RR186 pKa = 11.84YY187 pKa = 8.8RR188 pKa = 11.84QVPSDD193 pKa = 3.86VTCSQPFQDD202 pKa = 4.24CAMRR206 pKa = 11.84TPPNQDD212 pKa = 2.69CFAISVHH219 pKa = 5.68SLYY222 pKa = 10.65DD223 pKa = 3.34IPVDD227 pKa = 3.75EE228 pKa = 5.53LGCALLRR235 pKa = 11.84KK236 pKa = 9.07NVKK239 pKa = 9.15ILYY242 pKa = 9.05AALHH246 pKa = 5.77FSEE249 pKa = 5.72DD250 pKa = 3.8LLLGATDD257 pKa = 4.19APLGNIGARR266 pKa = 11.84FYY268 pKa = 11.36RR269 pKa = 11.84NGDD272 pKa = 3.24EE273 pKa = 4.23VTFGFDD279 pKa = 3.67GEE281 pKa = 4.6STLLYY286 pKa = 9.15THH288 pKa = 6.46SFKK291 pKa = 11.15NICGYY296 pKa = 7.53ITRR299 pKa = 11.84SFFYY303 pKa = 10.6AGAKK307 pKa = 7.48YY308 pKa = 10.44AYY310 pKa = 9.09MKK312 pKa = 10.55EE313 pKa = 4.12FMVKK317 pKa = 10.01RR318 pKa = 11.84VDD320 pKa = 3.44TVFCKK325 pKa = 10.42FVRR328 pKa = 11.84IDD330 pKa = 3.49TQCLYY335 pKa = 11.03KK336 pKa = 10.61SVFHH340 pKa = 5.82TQIDD344 pKa = 3.6SDD346 pKa = 4.19AVLEE350 pKa = 4.53GMDD353 pKa = 3.48SAFTARR359 pKa = 11.84RR360 pKa = 11.84QAAMLNAARR369 pKa = 11.84PLFKK373 pKa = 10.72DD374 pKa = 3.17KK375 pKa = 11.17AAFSVWFPNAIGKK388 pKa = 8.43VLIPIFRR395 pKa = 11.84GTTCLGAKK403 pKa = 9.55IKK405 pKa = 9.64MEE407 pKa = 3.89KK408 pKa = 10.27RR409 pKa = 11.84LVDD412 pKa = 3.45EE413 pKa = 4.89DD414 pKa = 3.97FVYY417 pKa = 10.39TILNHH422 pKa = 5.29IRR424 pKa = 11.84TYY426 pKa = 9.69TGKK429 pKa = 9.52QLSYY433 pKa = 10.83EE434 pKa = 4.14NVLSFVEE441 pKa = 4.76SIRR444 pKa = 11.84SRR446 pKa = 11.84VVINGANVRR455 pKa = 11.84SEE457 pKa = 3.84WNIPKK462 pKa = 10.14SDD464 pKa = 3.44IQDD467 pKa = 2.98IAMSLFLITKK477 pKa = 9.88LRR479 pKa = 11.84TLQDD483 pKa = 3.56SAVLSHH489 pKa = 7.2FDD491 pKa = 3.13IKK493 pKa = 11.08KK494 pKa = 10.37KK495 pKa = 10.94GFFDD499 pKa = 4.11CVRR502 pKa = 11.84EE503 pKa = 4.18TIMEE507 pKa = 4.13VTGSIFEE514 pKa = 4.82PLTQICIDD522 pKa = 3.94HH523 pKa = 6.46GWFKK527 pKa = 10.83IKK529 pKa = 10.06EE530 pKa = 4.19DD531 pKa = 3.52KK532 pKa = 11.21LKK534 pKa = 11.3VEE536 pKa = 4.51VPDD539 pKa = 3.56TSRR542 pKa = 11.84TFMSYY547 pKa = 8.14MQTEE551 pKa = 4.64FSEE554 pKa = 4.67SKK556 pKa = 10.63AIEE559 pKa = 4.32SLDD562 pKa = 3.07LWEE565 pKa = 5.68YY566 pKa = 11.16LDD568 pKa = 5.44QSNQLYY574 pKa = 7.8THH576 pKa = 6.26VSKK579 pKa = 11.03LMEE582 pKa = 5.03RR583 pKa = 11.84YY584 pKa = 9.49SLPDD588 pKa = 3.42FDD590 pKa = 4.71VEE592 pKa = 4.97KK593 pKa = 11.03FKK595 pKa = 11.05DD596 pKa = 3.46LCAHH600 pKa = 6.62LKK602 pKa = 10.17VGPEE606 pKa = 4.0VITAVIEE613 pKa = 4.07AVMDD617 pKa = 3.62KK618 pKa = 11.02SLGFTVVGGKK628 pKa = 9.86AEE630 pKa = 4.1HH631 pKa = 6.32QFTDD635 pKa = 3.13EE636 pKa = 3.98MKK638 pKa = 10.68AAVAVSADD646 pKa = 3.86YY647 pKa = 10.57GQHH650 pKa = 5.59CTDD653 pKa = 3.49KK654 pKa = 11.01KK655 pKa = 10.33QVASAACKK663 pKa = 10.76NEE665 pKa = 3.67VTKK668 pKa = 10.18PLKK671 pKa = 9.97KK672 pKa = 9.95QWVEE676 pKa = 3.68KK677 pKa = 7.99TTAALPLQATTEE689 pKa = 4.3KK690 pKa = 9.46TQHH693 pKa = 5.08FHH695 pKa = 7.43FNDD698 pKa = 3.4EE699 pKa = 4.51DD700 pKa = 3.92EE701 pKa = 5.12SISVEE706 pKa = 3.96NLHH709 pKa = 6.87LKK711 pKa = 10.39RR712 pKa = 11.84ADD714 pKa = 3.29TFKK717 pKa = 11.05ASKK720 pKa = 10.92ACMLYY725 pKa = 9.49TGTVRR730 pKa = 11.84EE731 pKa = 4.01QQMKK735 pKa = 10.79NFLDD739 pKa = 3.7YY740 pKa = 11.22LSASICATISNIEE753 pKa = 4.28KK754 pKa = 10.37VLSDD758 pKa = 3.27YY759 pKa = 9.97WVSGTQTYY767 pKa = 7.2QTYY770 pKa = 10.94GLWDD774 pKa = 3.78CKK776 pKa = 10.14KK777 pKa = 10.28KK778 pKa = 10.49KK779 pKa = 9.12WVLQPPTAGHH789 pKa = 5.64SWGIASIKK797 pKa = 10.26GEE799 pKa = 4.24HH800 pKa = 6.76FIVMLSFNEE809 pKa = 4.14NVEE812 pKa = 4.65PICDD816 pKa = 3.87SSWDD820 pKa = 3.9MVCVSNDD827 pKa = 3.19TKK829 pKa = 11.02LFSAMKK835 pKa = 9.65ILEE838 pKa = 4.52NLTPLPVKK846 pKa = 10.22EE847 pKa = 4.43SNAKK851 pKa = 7.91ITLVDD856 pKa = 4.08GVPGCGKK863 pKa = 8.25TAEE866 pKa = 4.39ILAKK870 pKa = 10.84ANFKK874 pKa = 10.59KK875 pKa = 10.67DD876 pKa = 3.98LILTQGKK883 pKa = 8.53QAAQMIRR890 pKa = 11.84KK891 pKa = 8.52RR892 pKa = 11.84ANAIDD897 pKa = 3.78CTKK900 pKa = 10.2PANTSNVRR908 pKa = 11.84TVDD911 pKa = 3.54SFLMNPKK918 pKa = 9.83PFTYY922 pKa = 8.69DD923 pKa = 3.26TLWIDD928 pKa = 3.74EE929 pKa = 4.32GLMLHH934 pKa = 6.37TGMVNYY940 pKa = 10.12CVLLSHH946 pKa = 6.35CTKK949 pKa = 10.59CFIYY953 pKa = 10.7GDD955 pKa = 3.79TQQIPFINRR964 pKa = 11.84VMNFDD969 pKa = 3.77YY970 pKa = 10.49PGHH973 pKa = 6.82LRR975 pKa = 11.84TIVVDD980 pKa = 3.55DD981 pKa = 3.81TEE983 pKa = 4.24KK984 pKa = 10.82RR985 pKa = 11.84RR986 pKa = 11.84ITSRR990 pKa = 11.84CPLDD994 pKa = 3.19VTFYY998 pKa = 10.03LTQRR1002 pKa = 11.84YY1003 pKa = 8.2KK1004 pKa = 11.04GPVMSTSSVTRR1015 pKa = 11.84SVEE1018 pKa = 4.18TKK1020 pKa = 10.34FVAGPARR1027 pKa = 11.84FEE1029 pKa = 4.15PRR1031 pKa = 11.84ATPLPGKK1038 pKa = 10.06IVTFTQADD1046 pKa = 3.86KK1047 pKa = 10.14EE1048 pKa = 4.58TLKK1051 pKa = 11.04KK1052 pKa = 10.25KK1053 pKa = 10.45GYY1055 pKa = 9.04NDD1057 pKa = 2.88VHH1059 pKa = 5.93TVHH1062 pKa = 7.38EE1063 pKa = 4.44IQGEE1067 pKa = 4.47TFNEE1071 pKa = 4.09VSLVRR1076 pKa = 11.84LTATPLNIISPEE1088 pKa = 4.11SPHH1091 pKa = 7.52VLVGLSRR1098 pKa = 11.84HH1099 pKa = 5.26TNRR1102 pKa = 11.84LTYY1105 pKa = 8.44YY1106 pKa = 9.69TVVHH1110 pKa = 6.72DD1111 pKa = 5.38CITYY1115 pKa = 10.33SISEE1119 pKa = 4.34LSRR1122 pKa = 11.84LSDD1125 pKa = 4.15YY1126 pKa = 11.22IFDD1129 pKa = 3.84IYY1131 pKa = 11.17SVEE1134 pKa = 4.08GQKK1137 pKa = 10.71AA1138 pKa = 2.99
MM1 pKa = 6.62TTSTTDD7 pKa = 2.67IKK9 pKa = 10.91QQITNVRR16 pKa = 11.84DD17 pKa = 3.73DD18 pKa = 4.12AAHH21 pKa = 6.6GAASLIKK28 pKa = 10.28GLATRR33 pKa = 11.84RR34 pKa = 11.84VYY36 pKa = 11.08DD37 pKa = 3.58EE38 pKa = 4.64AVNTLTLLDD47 pKa = 4.25KK48 pKa = 10.73RR49 pKa = 11.84PKK51 pKa = 10.21FSFSNLVSTEE61 pKa = 3.67NAKK64 pKa = 9.42MVTEE68 pKa = 5.5AYY70 pKa = 9.73PEE72 pKa = 4.04FNISFVGNRR81 pKa = 11.84NTVHH85 pKa = 6.58SLAGGLRR92 pKa = 11.84KK93 pKa = 9.93LEE95 pKa = 4.02MEE97 pKa = 4.48YY98 pKa = 11.29LMTLVPYY105 pKa = 10.68GSATFDD111 pKa = 2.71IGGNYY116 pKa = 8.98AQHH119 pKa = 6.51LLKK122 pKa = 10.75GRR124 pKa = 11.84AYY126 pKa = 8.8VHH128 pKa = 6.65CCNPCLDD135 pKa = 3.98FRR137 pKa = 11.84DD138 pKa = 3.57IARR141 pKa = 11.84NEE143 pKa = 4.27SYY145 pKa = 11.08KK146 pKa = 10.53DD147 pKa = 3.53TVDD150 pKa = 3.8SYY152 pKa = 11.66LKK154 pKa = 10.69RR155 pKa = 11.84FTTPDD160 pKa = 3.61CGWFSGAVRR169 pKa = 11.84AITARR174 pKa = 11.84PLPSFQKK181 pKa = 10.5EE182 pKa = 3.52AFEE185 pKa = 5.24RR186 pKa = 11.84YY187 pKa = 8.8RR188 pKa = 11.84QVPSDD193 pKa = 3.86VTCSQPFQDD202 pKa = 4.24CAMRR206 pKa = 11.84TPPNQDD212 pKa = 2.69CFAISVHH219 pKa = 5.68SLYY222 pKa = 10.65DD223 pKa = 3.34IPVDD227 pKa = 3.75EE228 pKa = 5.53LGCALLRR235 pKa = 11.84KK236 pKa = 9.07NVKK239 pKa = 9.15ILYY242 pKa = 9.05AALHH246 pKa = 5.77FSEE249 pKa = 5.72DD250 pKa = 3.8LLLGATDD257 pKa = 4.19APLGNIGARR266 pKa = 11.84FYY268 pKa = 11.36RR269 pKa = 11.84NGDD272 pKa = 3.24EE273 pKa = 4.23VTFGFDD279 pKa = 3.67GEE281 pKa = 4.6STLLYY286 pKa = 9.15THH288 pKa = 6.46SFKK291 pKa = 11.15NICGYY296 pKa = 7.53ITRR299 pKa = 11.84SFFYY303 pKa = 10.6AGAKK307 pKa = 7.48YY308 pKa = 10.44AYY310 pKa = 9.09MKK312 pKa = 10.55EE313 pKa = 4.12FMVKK317 pKa = 10.01RR318 pKa = 11.84VDD320 pKa = 3.44TVFCKK325 pKa = 10.42FVRR328 pKa = 11.84IDD330 pKa = 3.49TQCLYY335 pKa = 11.03KK336 pKa = 10.61SVFHH340 pKa = 5.82TQIDD344 pKa = 3.6SDD346 pKa = 4.19AVLEE350 pKa = 4.53GMDD353 pKa = 3.48SAFTARR359 pKa = 11.84RR360 pKa = 11.84QAAMLNAARR369 pKa = 11.84PLFKK373 pKa = 10.72DD374 pKa = 3.17KK375 pKa = 11.17AAFSVWFPNAIGKK388 pKa = 8.43VLIPIFRR395 pKa = 11.84GTTCLGAKK403 pKa = 9.55IKK405 pKa = 9.64MEE407 pKa = 3.89KK408 pKa = 10.27RR409 pKa = 11.84LVDD412 pKa = 3.45EE413 pKa = 4.89DD414 pKa = 3.97FVYY417 pKa = 10.39TILNHH422 pKa = 5.29IRR424 pKa = 11.84TYY426 pKa = 9.69TGKK429 pKa = 9.52QLSYY433 pKa = 10.83EE434 pKa = 4.14NVLSFVEE441 pKa = 4.76SIRR444 pKa = 11.84SRR446 pKa = 11.84VVINGANVRR455 pKa = 11.84SEE457 pKa = 3.84WNIPKK462 pKa = 10.14SDD464 pKa = 3.44IQDD467 pKa = 2.98IAMSLFLITKK477 pKa = 9.88LRR479 pKa = 11.84TLQDD483 pKa = 3.56SAVLSHH489 pKa = 7.2FDD491 pKa = 3.13IKK493 pKa = 11.08KK494 pKa = 10.37KK495 pKa = 10.94GFFDD499 pKa = 4.11CVRR502 pKa = 11.84EE503 pKa = 4.18TIMEE507 pKa = 4.13VTGSIFEE514 pKa = 4.82PLTQICIDD522 pKa = 3.94HH523 pKa = 6.46GWFKK527 pKa = 10.83IKK529 pKa = 10.06EE530 pKa = 4.19DD531 pKa = 3.52KK532 pKa = 11.21LKK534 pKa = 11.3VEE536 pKa = 4.51VPDD539 pKa = 3.56TSRR542 pKa = 11.84TFMSYY547 pKa = 8.14MQTEE551 pKa = 4.64FSEE554 pKa = 4.67SKK556 pKa = 10.63AIEE559 pKa = 4.32SLDD562 pKa = 3.07LWEE565 pKa = 5.68YY566 pKa = 11.16LDD568 pKa = 5.44QSNQLYY574 pKa = 7.8THH576 pKa = 6.26VSKK579 pKa = 11.03LMEE582 pKa = 5.03RR583 pKa = 11.84YY584 pKa = 9.49SLPDD588 pKa = 3.42FDD590 pKa = 4.71VEE592 pKa = 4.97KK593 pKa = 11.03FKK595 pKa = 11.05DD596 pKa = 3.46LCAHH600 pKa = 6.62LKK602 pKa = 10.17VGPEE606 pKa = 4.0VITAVIEE613 pKa = 4.07AVMDD617 pKa = 3.62KK618 pKa = 11.02SLGFTVVGGKK628 pKa = 9.86AEE630 pKa = 4.1HH631 pKa = 6.32QFTDD635 pKa = 3.13EE636 pKa = 3.98MKK638 pKa = 10.68AAVAVSADD646 pKa = 3.86YY647 pKa = 10.57GQHH650 pKa = 5.59CTDD653 pKa = 3.49KK654 pKa = 11.01KK655 pKa = 10.33QVASAACKK663 pKa = 10.76NEE665 pKa = 3.67VTKK668 pKa = 10.18PLKK671 pKa = 9.97KK672 pKa = 9.95QWVEE676 pKa = 3.68KK677 pKa = 7.99TTAALPLQATTEE689 pKa = 4.3KK690 pKa = 9.46TQHH693 pKa = 5.08FHH695 pKa = 7.43FNDD698 pKa = 3.4EE699 pKa = 4.51DD700 pKa = 3.92EE701 pKa = 5.12SISVEE706 pKa = 3.96NLHH709 pKa = 6.87LKK711 pKa = 10.39RR712 pKa = 11.84ADD714 pKa = 3.29TFKK717 pKa = 11.05ASKK720 pKa = 10.92ACMLYY725 pKa = 9.49TGTVRR730 pKa = 11.84EE731 pKa = 4.01QQMKK735 pKa = 10.79NFLDD739 pKa = 3.7YY740 pKa = 11.22LSASICATISNIEE753 pKa = 4.28KK754 pKa = 10.37VLSDD758 pKa = 3.27YY759 pKa = 9.97WVSGTQTYY767 pKa = 7.2QTYY770 pKa = 10.94GLWDD774 pKa = 3.78CKK776 pKa = 10.14KK777 pKa = 10.28KK778 pKa = 10.49KK779 pKa = 9.12WVLQPPTAGHH789 pKa = 5.64SWGIASIKK797 pKa = 10.26GEE799 pKa = 4.24HH800 pKa = 6.76FIVMLSFNEE809 pKa = 4.14NVEE812 pKa = 4.65PICDD816 pKa = 3.87SSWDD820 pKa = 3.9MVCVSNDD827 pKa = 3.19TKK829 pKa = 11.02LFSAMKK835 pKa = 9.65ILEE838 pKa = 4.52NLTPLPVKK846 pKa = 10.22EE847 pKa = 4.43SNAKK851 pKa = 7.91ITLVDD856 pKa = 4.08GVPGCGKK863 pKa = 8.25TAEE866 pKa = 4.39ILAKK870 pKa = 10.84ANFKK874 pKa = 10.59KK875 pKa = 10.67DD876 pKa = 3.98LILTQGKK883 pKa = 8.53QAAQMIRR890 pKa = 11.84KK891 pKa = 8.52RR892 pKa = 11.84ANAIDD897 pKa = 3.78CTKK900 pKa = 10.2PANTSNVRR908 pKa = 11.84TVDD911 pKa = 3.54SFLMNPKK918 pKa = 9.83PFTYY922 pKa = 8.69DD923 pKa = 3.26TLWIDD928 pKa = 3.74EE929 pKa = 4.32GLMLHH934 pKa = 6.37TGMVNYY940 pKa = 10.12CVLLSHH946 pKa = 6.35CTKK949 pKa = 10.59CFIYY953 pKa = 10.7GDD955 pKa = 3.79TQQIPFINRR964 pKa = 11.84VMNFDD969 pKa = 3.77YY970 pKa = 10.49PGHH973 pKa = 6.82LRR975 pKa = 11.84TIVVDD980 pKa = 3.55DD981 pKa = 3.81TEE983 pKa = 4.24KK984 pKa = 10.82RR985 pKa = 11.84RR986 pKa = 11.84ITSRR990 pKa = 11.84CPLDD994 pKa = 3.19VTFYY998 pKa = 10.03LTQRR1002 pKa = 11.84YY1003 pKa = 8.2KK1004 pKa = 11.04GPVMSTSSVTRR1015 pKa = 11.84SVEE1018 pKa = 4.18TKK1020 pKa = 10.34FVAGPARR1027 pKa = 11.84FEE1029 pKa = 4.15PRR1031 pKa = 11.84ATPLPGKK1038 pKa = 10.06IVTFTQADD1046 pKa = 3.86KK1047 pKa = 10.14EE1048 pKa = 4.58TLKK1051 pKa = 11.04KK1052 pKa = 10.25KK1053 pKa = 10.45GYY1055 pKa = 9.04NDD1057 pKa = 2.88VHH1059 pKa = 5.93TVHH1062 pKa = 7.38EE1063 pKa = 4.44IQGEE1067 pKa = 4.47TFNEE1071 pKa = 4.09VSLVRR1076 pKa = 11.84LTATPLNIISPEE1088 pKa = 4.11SPHH1091 pKa = 7.52VLVGLSRR1098 pKa = 11.84HH1099 pKa = 5.26TNRR1102 pKa = 11.84LTYY1105 pKa = 8.44YY1106 pKa = 9.69TVVHH1110 pKa = 6.72DD1111 pKa = 5.38CITYY1115 pKa = 10.33SISEE1119 pKa = 4.34LSRR1122 pKa = 11.84LSDD1125 pKa = 4.15YY1126 pKa = 11.22IFDD1129 pKa = 3.84IYY1131 pKa = 11.17SVEE1134 pKa = 4.08GQKK1137 pKa = 10.71AA1138 pKa = 2.99
Molecular weight: 128.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3226 |
158 |
1646 |
806.5 |
90.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.789 ± 0.412 | 2.356 ± 0.348 |
6.603 ± 0.267 | 5.053 ± 0.146 |
5.332 ± 0.415 | 5.115 ± 0.242 |
2.294 ± 0.208 | 6.045 ± 0.228 |
7.347 ± 0.559 | 8.586 ± 0.392 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.232 ± 0.223 | 4.371 ± 0.243 |
3.72 ± 0.154 | 3.565 ± 0.2 |
4.278 ± 0.204 | 6.913 ± 0.145 |
7.44 ± 0.541 | 7.161 ± 0.277 |
1.085 ± 0.142 | 3.689 ± 0.311 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
