
Blastococcus sp. CT_GayMR16
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus; unclassified Blastococcus
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
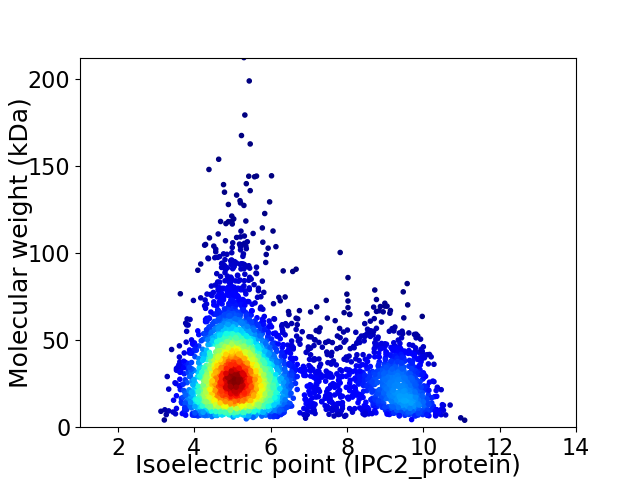
Virtual 2D-PAGE plot for 4365 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y9QC28|A0A4Y9QC28_9ACTN 30S ribosomal protein S14 type Z OS=Blastococcus sp. CT_GayMR16 OX=2559607 GN=rpsZ PE=3 SV=1
MM1 pKa = 7.98RR2 pKa = 11.84EE3 pKa = 3.97ALHH6 pKa = 6.43VKK8 pKa = 10.0LSKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.78SSLVATAIAAAMLMTACGGSDD34 pKa = 3.48DD35 pKa = 5.12SGDD38 pKa = 3.45GGAASDD44 pKa = 3.89GGSFSIYY51 pKa = 9.67IGEE54 pKa = 4.27PEE56 pKa = 4.07NPIVPGNTSEE66 pKa = 4.82SEE68 pKa = 4.06GSQVVEE74 pKa = 4.27ALWTGLVQYY83 pKa = 11.03ADD85 pKa = 4.31DD86 pKa = 5.04SSVQYY91 pKa = 10.34TGVAEE96 pKa = 4.49SIEE99 pKa = 4.38SDD101 pKa = 4.5DD102 pKa = 3.6ATTWTVKK109 pKa = 10.8LKK111 pKa = 10.87DD112 pKa = 2.95GWTFHH117 pKa = 7.56DD118 pKa = 4.4GTPVTAASFVDD129 pKa = 3.44AWNYY133 pKa = 7.46TALSTNAQGGSYY145 pKa = 10.03FFANVEE151 pKa = 4.45GYY153 pKa = 9.75DD154 pKa = 3.63ALQAEE159 pKa = 4.71TDD161 pKa = 3.57DD162 pKa = 4.93DD163 pKa = 5.35GNVITPPAAEE173 pKa = 4.22EE174 pKa = 3.99MTGLEE179 pKa = 4.26VVDD182 pKa = 4.51DD183 pKa = 3.75QTFTVTLTAPFAQYY197 pKa = 10.45PVTVGYY203 pKa = 10.18NAFYY207 pKa = 9.88PLPEE211 pKa = 4.51SFFEE215 pKa = 4.58DD216 pKa = 3.92PEE218 pKa = 4.66AAGKK222 pKa = 8.13TPVGNGPFKK231 pKa = 10.99ADD233 pKa = 3.35GEE235 pKa = 4.49FVPGQGFTLSRR246 pKa = 11.84YY247 pKa = 9.99DD248 pKa = 4.06DD249 pKa = 3.96YY250 pKa = 11.91AGEE253 pKa = 4.56DD254 pKa = 3.31AAQADD259 pKa = 4.04SVEE262 pKa = 4.46FKK264 pKa = 11.02VYY266 pKa = 10.6TDD268 pKa = 3.19INTAYY273 pKa = 9.8TDD275 pKa = 3.75TQGGNLDD282 pKa = 3.87ILNNIPPDD290 pKa = 4.8AISSAPDD297 pKa = 3.3EE298 pKa = 4.69FGDD301 pKa = 4.54RR302 pKa = 11.84YY303 pKa = 11.08GEE305 pKa = 4.29TPSSSFTYY313 pKa = 9.78MGFPTYY319 pKa = 10.55DD320 pKa = 2.96PRR322 pKa = 11.84YY323 pKa = 7.51ADD325 pKa = 3.09KK326 pKa = 10.68RR327 pKa = 11.84VRR329 pKa = 11.84QAFSLAIDD337 pKa = 3.79RR338 pKa = 11.84EE339 pKa = 4.6AITEE343 pKa = 4.51AIFNGTRR350 pKa = 11.84APAFSVIAPVVDD362 pKa = 4.57GSRR365 pKa = 11.84DD366 pKa = 3.65DD367 pKa = 3.67ACQYY371 pKa = 10.09CQVDD375 pKa = 3.54VEE377 pKa = 4.55RR378 pKa = 11.84ANEE381 pKa = 4.0LLDD384 pKa = 3.51EE385 pKa = 5.03AGFDD389 pKa = 3.36RR390 pKa = 11.84TQPVDD395 pKa = 2.84LWFNAGAGHH404 pKa = 6.64DD405 pKa = 3.6AWVEE409 pKa = 3.78AVGNQLRR416 pKa = 11.84EE417 pKa = 3.96NLGVEE422 pKa = 4.65FTLQGNLDD430 pKa = 3.61FSEE433 pKa = 4.17YY434 pKa = 10.96LPLGDD439 pKa = 3.69AKK441 pKa = 11.08GYY443 pKa = 8.02TGPFRR448 pKa = 11.84LGWSMDD454 pKa = 3.44YY455 pKa = 10.51PSPQNYY461 pKa = 9.97LEE463 pKa = 4.5PLYY466 pKa = 9.99STQAQPPAGSNSAFYY481 pKa = 11.16SNPEE485 pKa = 3.3FDD487 pKa = 4.29ALVAEE492 pKa = 5.16GNAASSNDD500 pKa = 3.51EE501 pKa = 5.34AIDD504 pKa = 4.58LYY506 pKa = 11.01QQAEE510 pKa = 4.21DD511 pKa = 4.84LLLEE515 pKa = 4.96DD516 pKa = 4.41MPIMPMFFGLEE527 pKa = 3.66QTVWSEE533 pKa = 3.78NVSDD537 pKa = 3.89VKK539 pKa = 11.2VDD541 pKa = 2.69IFGRR545 pKa = 11.84VNVASVTVKK554 pKa = 10.8
MM1 pKa = 7.98RR2 pKa = 11.84EE3 pKa = 3.97ALHH6 pKa = 6.43VKK8 pKa = 10.0LSKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.78SSLVATAIAAAMLMTACGGSDD34 pKa = 3.48DD35 pKa = 5.12SGDD38 pKa = 3.45GGAASDD44 pKa = 3.89GGSFSIYY51 pKa = 9.67IGEE54 pKa = 4.27PEE56 pKa = 4.07NPIVPGNTSEE66 pKa = 4.82SEE68 pKa = 4.06GSQVVEE74 pKa = 4.27ALWTGLVQYY83 pKa = 11.03ADD85 pKa = 4.31DD86 pKa = 5.04SSVQYY91 pKa = 10.34TGVAEE96 pKa = 4.49SIEE99 pKa = 4.38SDD101 pKa = 4.5DD102 pKa = 3.6ATTWTVKK109 pKa = 10.8LKK111 pKa = 10.87DD112 pKa = 2.95GWTFHH117 pKa = 7.56DD118 pKa = 4.4GTPVTAASFVDD129 pKa = 3.44AWNYY133 pKa = 7.46TALSTNAQGGSYY145 pKa = 10.03FFANVEE151 pKa = 4.45GYY153 pKa = 9.75DD154 pKa = 3.63ALQAEE159 pKa = 4.71TDD161 pKa = 3.57DD162 pKa = 4.93DD163 pKa = 5.35GNVITPPAAEE173 pKa = 4.22EE174 pKa = 3.99MTGLEE179 pKa = 4.26VVDD182 pKa = 4.51DD183 pKa = 3.75QTFTVTLTAPFAQYY197 pKa = 10.45PVTVGYY203 pKa = 10.18NAFYY207 pKa = 9.88PLPEE211 pKa = 4.51SFFEE215 pKa = 4.58DD216 pKa = 3.92PEE218 pKa = 4.66AAGKK222 pKa = 8.13TPVGNGPFKK231 pKa = 10.99ADD233 pKa = 3.35GEE235 pKa = 4.49FVPGQGFTLSRR246 pKa = 11.84YY247 pKa = 9.99DD248 pKa = 4.06DD249 pKa = 3.96YY250 pKa = 11.91AGEE253 pKa = 4.56DD254 pKa = 3.31AAQADD259 pKa = 4.04SVEE262 pKa = 4.46FKK264 pKa = 11.02VYY266 pKa = 10.6TDD268 pKa = 3.19INTAYY273 pKa = 9.8TDD275 pKa = 3.75TQGGNLDD282 pKa = 3.87ILNNIPPDD290 pKa = 4.8AISSAPDD297 pKa = 3.3EE298 pKa = 4.69FGDD301 pKa = 4.54RR302 pKa = 11.84YY303 pKa = 11.08GEE305 pKa = 4.29TPSSSFTYY313 pKa = 9.78MGFPTYY319 pKa = 10.55DD320 pKa = 2.96PRR322 pKa = 11.84YY323 pKa = 7.51ADD325 pKa = 3.09KK326 pKa = 10.68RR327 pKa = 11.84VRR329 pKa = 11.84QAFSLAIDD337 pKa = 3.79RR338 pKa = 11.84EE339 pKa = 4.6AITEE343 pKa = 4.51AIFNGTRR350 pKa = 11.84APAFSVIAPVVDD362 pKa = 4.57GSRR365 pKa = 11.84DD366 pKa = 3.65DD367 pKa = 3.67ACQYY371 pKa = 10.09CQVDD375 pKa = 3.54VEE377 pKa = 4.55RR378 pKa = 11.84ANEE381 pKa = 4.0LLDD384 pKa = 3.51EE385 pKa = 5.03AGFDD389 pKa = 3.36RR390 pKa = 11.84TQPVDD395 pKa = 2.84LWFNAGAGHH404 pKa = 6.64DD405 pKa = 3.6AWVEE409 pKa = 3.78AVGNQLRR416 pKa = 11.84EE417 pKa = 3.96NLGVEE422 pKa = 4.65FTLQGNLDD430 pKa = 3.61FSEE433 pKa = 4.17YY434 pKa = 10.96LPLGDD439 pKa = 3.69AKK441 pKa = 11.08GYY443 pKa = 8.02TGPFRR448 pKa = 11.84LGWSMDD454 pKa = 3.44YY455 pKa = 10.51PSPQNYY461 pKa = 9.97LEE463 pKa = 4.5PLYY466 pKa = 9.99STQAQPPAGSNSAFYY481 pKa = 11.16SNPEE485 pKa = 3.3FDD487 pKa = 4.29ALVAEE492 pKa = 5.16GNAASSNDD500 pKa = 3.51EE501 pKa = 5.34AIDD504 pKa = 4.58LYY506 pKa = 11.01QQAEE510 pKa = 4.21DD511 pKa = 4.84LLLEE515 pKa = 4.96DD516 pKa = 4.41MPIMPMFFGLEE527 pKa = 3.66QTVWSEE533 pKa = 3.78NVSDD537 pKa = 3.89VKK539 pKa = 11.2VDD541 pKa = 2.69IFGRR545 pKa = 11.84VNVASVTVKK554 pKa = 10.8
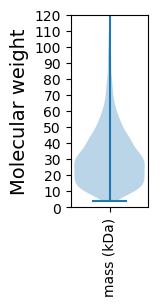
Molecular weight: 59.72 kDa
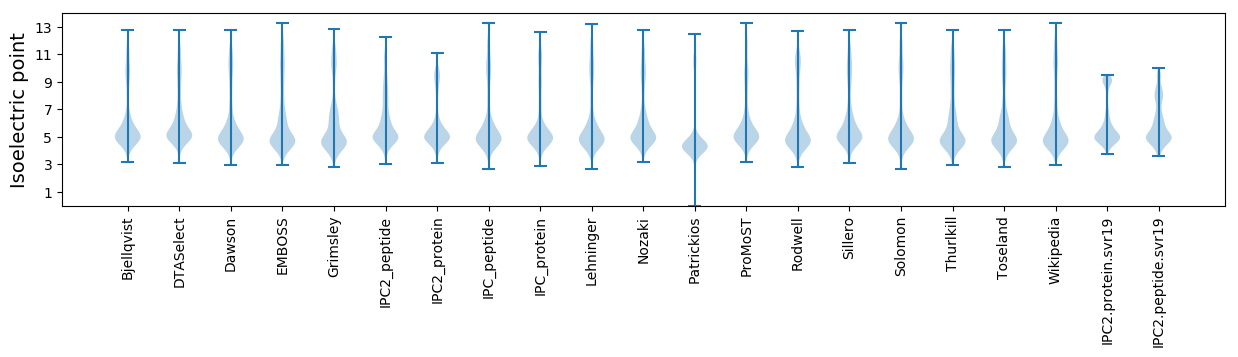
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y9PXA5|A0A4Y9PXA5_9ACTN ANTAR domain-containing protein OS=Blastococcus sp. CT_GayMR16 OX=2559607 GN=E4P38_20755 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
Molecular weight: 4.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1348885 |
32 |
1976 |
309.0 |
32.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.88 ± 0.059 | 0.677 ± 0.01 |
6.372 ± 0.03 | 5.602 ± 0.038 |
2.67 ± 0.021 | 9.499 ± 0.038 |
1.975 ± 0.018 | 3.196 ± 0.026 |
1.578 ± 0.025 | 10.601 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.699 ± 0.013 | 1.618 ± 0.02 |
6.059 ± 0.03 | 2.77 ± 0.02 |
7.87 ± 0.047 | 5.142 ± 0.026 |
6.022 ± 0.031 | 9.497 ± 0.033 |
1.485 ± 0.016 | 1.789 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
