
Xylanimonas oleitrophica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Promicromonosporaceae; Xylanimonas
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
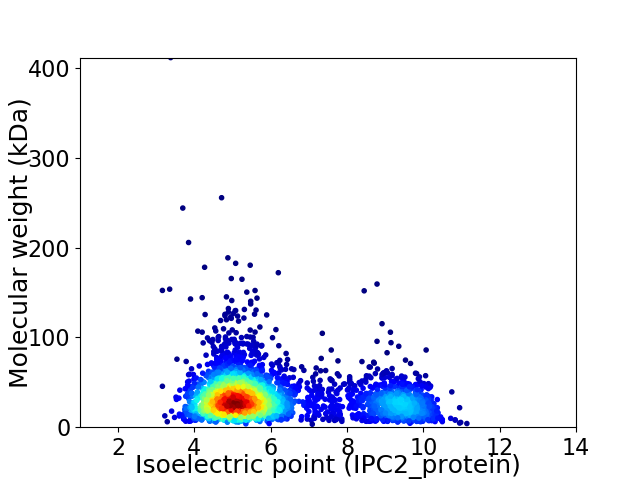
Virtual 2D-PAGE plot for 3108 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2W5WXY7|A0A2W5WXY7_9MICO Uncharacterized N-acetyltransferase DNL40_02185 OS=Xylanimonas oleitrophica OX=2607479 GN=DNL40_02185 PE=3 SV=1
MM1 pKa = 7.43IRR3 pKa = 11.84RR4 pKa = 11.84HH5 pKa = 5.3SAAKK9 pKa = 10.28GLALAAALGLGLAACSTSAEE29 pKa = 4.22GSGGEE34 pKa = 4.38GSDD37 pKa = 3.33SAEE40 pKa = 3.83PLVIGTILPITGTLAFLGPPEE61 pKa = 4.03VAGVGLAVDD70 pKa = 5.78DD71 pKa = 4.17INEE74 pKa = 4.31AGGVLGNDD82 pKa = 3.83VSVEE86 pKa = 3.86WGDD89 pKa = 4.39SGDD92 pKa = 3.79TTDD95 pKa = 4.95LSVAGGTATDD105 pKa = 5.24LIGKK109 pKa = 8.41GVSAVVGAASSGVSLAVVDD128 pKa = 4.01QFEE131 pKa = 4.48EE132 pKa = 4.36AGVMQISPANTATDD146 pKa = 3.94LSGYY150 pKa = 10.78GDD152 pKa = 4.22FFARR156 pKa = 11.84TAPPDD161 pKa = 4.04TVQGAALGSQILDD174 pKa = 3.87DD175 pKa = 3.5GHH177 pKa = 8.15LRR179 pKa = 11.84VGFLVQNEE187 pKa = 4.41AYY189 pKa = 10.61GNGLRR194 pKa = 11.84DD195 pKa = 3.76NVQKK199 pKa = 10.83AIEE202 pKa = 4.43SGGGEE207 pKa = 4.02AVYY210 pKa = 10.24GATGAGQEE218 pKa = 4.22FAPGEE223 pKa = 4.51TNFGAQVTALLATQPDD239 pKa = 4.88AIAIIAFEE247 pKa = 4.15EE248 pKa = 4.32TVAIINEE255 pKa = 4.31LVAQGWSFSGTTYY268 pKa = 10.59FCDD271 pKa = 4.05GNLSNYY277 pKa = 9.6GDD279 pKa = 3.62QFDD282 pKa = 4.38AGTLAGVKK290 pKa = 8.92GTLPGAQADD299 pKa = 3.98EE300 pKa = 4.63EE301 pKa = 4.37FQQRR305 pKa = 11.84LNDD308 pKa = 3.49WYY310 pKa = 11.24SEE312 pKa = 4.23NEE314 pKa = 4.11SGTLNDD320 pKa = 3.14YY321 pKa = 11.13SYY323 pKa = 11.51AAEE326 pKa = 4.7SYY328 pKa = 10.76DD329 pKa = 3.61ATILAALAAVRR340 pKa = 11.84GGATDD345 pKa = 3.68GQTIADD351 pKa = 4.09NIKK354 pKa = 10.22AVSGAEE360 pKa = 3.88GGTEE364 pKa = 3.58VTTFAEE370 pKa = 4.7GVEE373 pKa = 3.95ALEE376 pKa = 4.75NGDD379 pKa = 3.92EE380 pKa = 3.72IHH382 pKa = 6.42YY383 pKa = 10.0VGKK386 pKa = 10.72AGIGPLNEE394 pKa = 4.58SNDD397 pKa = 3.5PSSAFIGIYY406 pKa = 10.16EE407 pKa = 4.2YY408 pKa = 11.52GDD410 pKa = 4.15DD411 pKa = 3.61NTYY414 pKa = 9.45TYY416 pKa = 11.0SRR418 pKa = 11.84SVEE421 pKa = 4.11GKK423 pKa = 9.68AA424 pKa = 4.1
MM1 pKa = 7.43IRR3 pKa = 11.84RR4 pKa = 11.84HH5 pKa = 5.3SAAKK9 pKa = 10.28GLALAAALGLGLAACSTSAEE29 pKa = 4.22GSGGEE34 pKa = 4.38GSDD37 pKa = 3.33SAEE40 pKa = 3.83PLVIGTILPITGTLAFLGPPEE61 pKa = 4.03VAGVGLAVDD70 pKa = 5.78DD71 pKa = 4.17INEE74 pKa = 4.31AGGVLGNDD82 pKa = 3.83VSVEE86 pKa = 3.86WGDD89 pKa = 4.39SGDD92 pKa = 3.79TTDD95 pKa = 4.95LSVAGGTATDD105 pKa = 5.24LIGKK109 pKa = 8.41GVSAVVGAASSGVSLAVVDD128 pKa = 4.01QFEE131 pKa = 4.48EE132 pKa = 4.36AGVMQISPANTATDD146 pKa = 3.94LSGYY150 pKa = 10.78GDD152 pKa = 4.22FFARR156 pKa = 11.84TAPPDD161 pKa = 4.04TVQGAALGSQILDD174 pKa = 3.87DD175 pKa = 3.5GHH177 pKa = 8.15LRR179 pKa = 11.84VGFLVQNEE187 pKa = 4.41AYY189 pKa = 10.61GNGLRR194 pKa = 11.84DD195 pKa = 3.76NVQKK199 pKa = 10.83AIEE202 pKa = 4.43SGGGEE207 pKa = 4.02AVYY210 pKa = 10.24GATGAGQEE218 pKa = 4.22FAPGEE223 pKa = 4.51TNFGAQVTALLATQPDD239 pKa = 4.88AIAIIAFEE247 pKa = 4.15EE248 pKa = 4.32TVAIINEE255 pKa = 4.31LVAQGWSFSGTTYY268 pKa = 10.59FCDD271 pKa = 4.05GNLSNYY277 pKa = 9.6GDD279 pKa = 3.62QFDD282 pKa = 4.38AGTLAGVKK290 pKa = 8.92GTLPGAQADD299 pKa = 3.98EE300 pKa = 4.63EE301 pKa = 4.37FQQRR305 pKa = 11.84LNDD308 pKa = 3.49WYY310 pKa = 11.24SEE312 pKa = 4.23NEE314 pKa = 4.11SGTLNDD320 pKa = 3.14YY321 pKa = 11.13SYY323 pKa = 11.51AAEE326 pKa = 4.7SYY328 pKa = 10.76DD329 pKa = 3.61ATILAALAAVRR340 pKa = 11.84GGATDD345 pKa = 3.68GQTIADD351 pKa = 4.09NIKK354 pKa = 10.22AVSGAEE360 pKa = 3.88GGTEE364 pKa = 3.58VTTFAEE370 pKa = 4.7GVEE373 pKa = 3.95ALEE376 pKa = 4.75NGDD379 pKa = 3.92EE380 pKa = 3.72IHH382 pKa = 6.42YY383 pKa = 10.0VGKK386 pKa = 10.72AGIGPLNEE394 pKa = 4.58SNDD397 pKa = 3.5PSSAFIGIYY406 pKa = 10.16EE407 pKa = 4.2YY408 pKa = 11.52GDD410 pKa = 4.15DD411 pKa = 3.61NTYY414 pKa = 9.45TYY416 pKa = 11.0SRR418 pKa = 11.84SVEE421 pKa = 4.11GKK423 pKa = 9.68AA424 pKa = 4.1
Molecular weight: 42.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2W5YCU3|A0A2W5YCU3_9MICO Alpha/beta hydrolase OS=Xylanimonas oleitrophica OX=2607479 GN=DNL40_13520 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1032422 |
30 |
4136 |
332.2 |
35.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.699 ± 0.075 | 0.521 ± 0.012 |
6.126 ± 0.044 | 5.687 ± 0.046 |
2.453 ± 0.029 | 9.648 ± 0.045 |
2.105 ± 0.027 | 2.635 ± 0.036 |
1.486 ± 0.03 | 10.154 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.578 ± 0.019 | 1.433 ± 0.026 |
6.106 ± 0.039 | 2.859 ± 0.025 |
7.879 ± 0.058 | 4.806 ± 0.029 |
6.207 ± 0.049 | 10.317 ± 0.053 |
1.517 ± 0.022 | 1.784 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
