
Cordyceps javanica
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales;
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
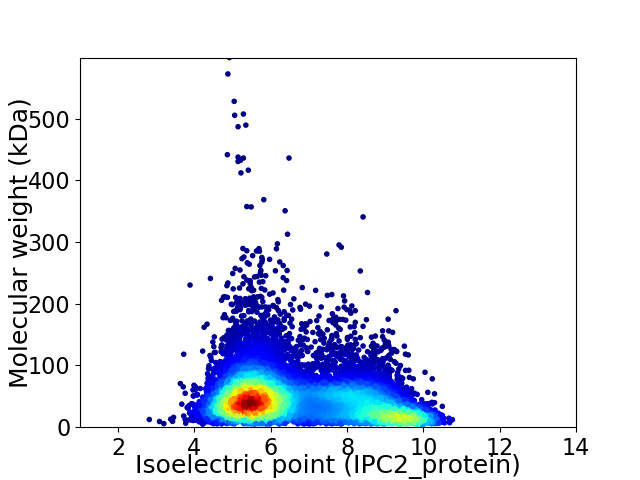
Virtual 2D-PAGE plot for 11436 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A545UUR3|A0A545UUR3_9HYPO Cytochrome P450 OS=Cordyceps javanica OX=43265 GN=IF1G_07785 PE=3 SV=1
MM1 pKa = 7.48LVNLSTIAALGAVASAYY18 pKa = 10.03DD19 pKa = 3.78LPDD22 pKa = 3.5NLRR25 pKa = 11.84SLYY28 pKa = 9.48QAHH31 pKa = 7.34LSGEE35 pKa = 4.36CGNALSGPFSTGAVYY50 pKa = 10.78CGDD53 pKa = 3.41IPNAIFLKK61 pKa = 10.82GPDD64 pKa = 3.74GEE66 pKa = 4.42YY67 pKa = 11.07DD68 pKa = 4.74NMDD71 pKa = 3.85IDD73 pKa = 5.68CDD75 pKa = 3.92GANNAAGDD83 pKa = 4.18CANDD87 pKa = 3.92PSGQGQTAFVDD98 pKa = 4.13TVSTYY103 pKa = 11.07GIRR106 pKa = 11.84DD107 pKa = 3.32LDD109 pKa = 4.51ANLHH113 pKa = 5.92SYY115 pKa = 10.12VVFGNEE121 pKa = 3.57GGSPSFSPRR130 pKa = 11.84DD131 pKa = 3.41HH132 pKa = 6.52GVRR135 pKa = 11.84PLSVMAVVCNNQFYY149 pKa = 10.56GVWGDD154 pKa = 3.78TNGFTSTGEE163 pKa = 3.81ASLALAKK170 pKa = 10.61LCFPNDD176 pKa = 3.86GLNGDD181 pKa = 4.43AGHH184 pKa = 7.3DD185 pKa = 3.88EE186 pKa = 4.36KK187 pKa = 11.46DD188 pKa = 3.49VLYY191 pKa = 10.81LAFTGDD197 pKa = 3.35EE198 pKa = 4.24AVPGRR203 pKa = 11.84DD204 pKa = 3.23GANWAAGSPEE214 pKa = 4.03EE215 pKa = 4.75FEE217 pKa = 5.86DD218 pKa = 3.96SIKK221 pKa = 10.99AIGDD225 pKa = 3.55RR226 pKa = 11.84LVASLPSS233 pKa = 3.31
MM1 pKa = 7.48LVNLSTIAALGAVASAYY18 pKa = 10.03DD19 pKa = 3.78LPDD22 pKa = 3.5NLRR25 pKa = 11.84SLYY28 pKa = 9.48QAHH31 pKa = 7.34LSGEE35 pKa = 4.36CGNALSGPFSTGAVYY50 pKa = 10.78CGDD53 pKa = 3.41IPNAIFLKK61 pKa = 10.82GPDD64 pKa = 3.74GEE66 pKa = 4.42YY67 pKa = 11.07DD68 pKa = 4.74NMDD71 pKa = 3.85IDD73 pKa = 5.68CDD75 pKa = 3.92GANNAAGDD83 pKa = 4.18CANDD87 pKa = 3.92PSGQGQTAFVDD98 pKa = 4.13TVSTYY103 pKa = 11.07GIRR106 pKa = 11.84DD107 pKa = 3.32LDD109 pKa = 4.51ANLHH113 pKa = 5.92SYY115 pKa = 10.12VVFGNEE121 pKa = 3.57GGSPSFSPRR130 pKa = 11.84DD131 pKa = 3.41HH132 pKa = 6.52GVRR135 pKa = 11.84PLSVMAVVCNNQFYY149 pKa = 10.56GVWGDD154 pKa = 3.78TNGFTSTGEE163 pKa = 3.81ASLALAKK170 pKa = 10.61LCFPNDD176 pKa = 3.86GLNGDD181 pKa = 4.43AGHH184 pKa = 7.3DD185 pKa = 3.88EE186 pKa = 4.36KK187 pKa = 11.46DD188 pKa = 3.49VLYY191 pKa = 10.81LAFTGDD197 pKa = 3.35EE198 pKa = 4.24AVPGRR203 pKa = 11.84DD204 pKa = 3.23GANWAAGSPEE214 pKa = 4.03EE215 pKa = 4.75FEE217 pKa = 5.86DD218 pKa = 3.96SIKK221 pKa = 10.99AIGDD225 pKa = 3.55RR226 pKa = 11.84LVASLPSS233 pKa = 3.31
Molecular weight: 24.17 kDa
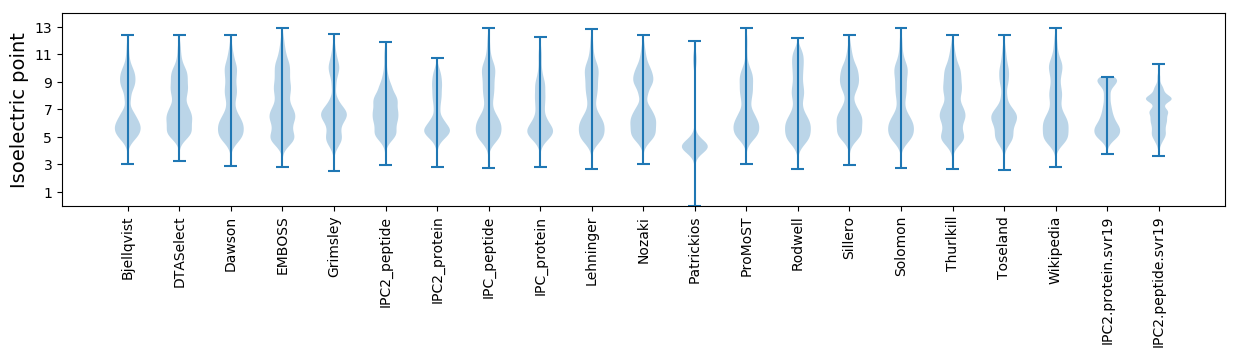
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A545UVV7|A0A545UVV7_9HYPO Uncharacterized protein OS=Cordyceps javanica OX=43265 GN=IF1G_07329 PE=4 SV=1
MM1 pKa = 7.57IGSTYY6 pKa = 11.11GNACLVRR13 pKa = 11.84FASALSEE20 pKa = 3.87RR21 pKa = 11.84KK22 pKa = 9.14RR23 pKa = 11.84RR24 pKa = 11.84KK25 pKa = 9.99KK26 pKa = 9.94IAQSPAAEE34 pKa = 4.48CGASRR39 pKa = 11.84WEE41 pKa = 4.47RR42 pKa = 11.84ICCLLTDD49 pKa = 4.07RR50 pKa = 11.84TEE52 pKa = 4.45SKK54 pKa = 10.36PPNDD58 pKa = 2.94GRR60 pKa = 11.84FGLRR64 pKa = 11.84ASAQTRR70 pKa = 11.84NSSTCVSGCFLGSRR84 pKa = 11.84NKK86 pKa = 10.2LLHH89 pKa = 6.85SLLQQHH95 pKa = 5.95ARR97 pKa = 11.84YY98 pKa = 9.37RR99 pKa = 3.69
MM1 pKa = 7.57IGSTYY6 pKa = 11.11GNACLVRR13 pKa = 11.84FASALSEE20 pKa = 3.87RR21 pKa = 11.84KK22 pKa = 9.14RR23 pKa = 11.84RR24 pKa = 11.84KK25 pKa = 9.99KK26 pKa = 9.94IAQSPAAEE34 pKa = 4.48CGASRR39 pKa = 11.84WEE41 pKa = 4.47RR42 pKa = 11.84ICCLLTDD49 pKa = 4.07RR50 pKa = 11.84TEE52 pKa = 4.45SKK54 pKa = 10.36PPNDD58 pKa = 2.94GRR60 pKa = 11.84FGLRR64 pKa = 11.84ASAQTRR70 pKa = 11.84NSSTCVSGCFLGSRR84 pKa = 11.84NKK86 pKa = 10.2LLHH89 pKa = 6.85SLLQQHH95 pKa = 5.95ARR97 pKa = 11.84YY98 pKa = 9.37RR99 pKa = 3.69
Molecular weight: 11.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5262576 |
35 |
5499 |
460.2 |
50.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.799 ± 0.024 | 1.321 ± 0.01 |
5.854 ± 0.019 | 5.768 ± 0.023 |
3.604 ± 0.015 | 7.151 ± 0.022 |
2.43 ± 0.011 | 4.513 ± 0.02 |
4.505 ± 0.021 | 8.858 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.226 ± 0.009 | 3.423 ± 0.014 |
5.936 ± 0.023 | 4.01 ± 0.017 |
6.407 ± 0.02 | 8.012 ± 0.025 |
5.847 ± 0.016 | 6.249 ± 0.018 |
1.449 ± 0.008 | 2.637 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
