
Ardenticatena maritima
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi;
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
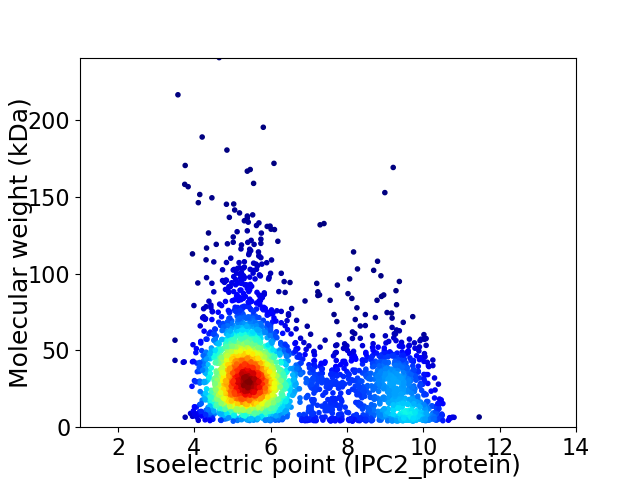
Virtual 2D-PAGE plot for 3144 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M8KAR0|A0A0M8KAR0_9CHLR Uncharacterized protein OS=Ardenticatena maritima OX=872965 GN=ARMA_2199 PE=3 SV=1
MM1 pKa = 7.25VLLLTFSLLVVFIFVPALWVTRR23 pKa = 11.84TTVAHH28 pKa = 6.96AAPEE32 pKa = 4.5GGALIVINEE41 pKa = 4.2FSVKK45 pKa = 9.23STDD48 pKa = 2.92WVEE51 pKa = 4.37LYY53 pKa = 8.48NTTAVTQSVAGWYY66 pKa = 10.45VSDD69 pKa = 4.34TGCGSPTTTMTGTIAPNDD87 pKa = 3.56FRR89 pKa = 11.84VLTVSDD95 pKa = 3.31AGGNFNFSSTAEE107 pKa = 4.22VISLCDD113 pKa = 3.4NAHH116 pKa = 6.01NEE118 pKa = 4.1VDD120 pKa = 3.52RR121 pKa = 11.84VAYY124 pKa = 10.0GIDD127 pKa = 3.31GGAPVPPSNGAFIYY141 pKa = 9.4STARR145 pKa = 11.84VADD148 pKa = 4.24GQDD151 pKa = 3.35TDD153 pKa = 4.99NDD155 pKa = 3.98AADD158 pKa = 3.83WNLDD162 pKa = 3.52STPTPGTPNDD172 pKa = 3.88APGTALGSSLLINEE186 pKa = 3.95IDD188 pKa = 3.86VYY190 pKa = 10.7PDD192 pKa = 3.18SGNDD196 pKa = 3.18IIEE199 pKa = 4.9LYY201 pKa = 10.96NPTTTPITITGWWVSDD217 pKa = 3.41GDD219 pKa = 3.75EE220 pKa = 4.21VAQVVGTVVVEE231 pKa = 4.31PGSVVALEE239 pKa = 4.33VGVDD243 pKa = 3.86LQDD246 pKa = 3.85EE247 pKa = 4.89GTTGLNFTSSDD258 pKa = 3.18VAYY261 pKa = 10.62LFDD264 pKa = 4.16EE265 pKa = 4.0NRR267 pKa = 11.84VRR269 pKa = 11.84VDD271 pKa = 3.09QLGWNGEE278 pKa = 4.11FEE280 pKa = 4.5NGCFARR286 pKa = 11.84VPDD289 pKa = 4.15GAGPNDD295 pKa = 4.26GYY297 pKa = 11.52DD298 pKa = 3.22WLSSGGGATLLDD310 pKa = 4.42LPCSLGAINGGPVVINEE327 pKa = 4.4FAPKK331 pKa = 7.81GTEE334 pKa = 3.53WVEE337 pKa = 3.83LYY339 pKa = 10.57NRR341 pKa = 11.84SPLTQTLTGWYY352 pKa = 8.32LTDD355 pKa = 3.8TACTGATSLITDD367 pKa = 4.47TTPIAPGGFFTVTNGAPGDD386 pKa = 3.87NFALSNNGDD395 pKa = 3.79TLLLCMADD403 pKa = 3.29NTLADD408 pKa = 3.51SVAYY412 pKa = 9.72GNSGGAPLASSTGAGDD428 pKa = 3.55ASIARR433 pKa = 11.84VSDD436 pKa = 3.76GVRR439 pKa = 11.84TGDD442 pKa = 5.69DD443 pKa = 3.33AADD446 pKa = 3.59WNVDD450 pKa = 3.28KK451 pKa = 10.93TPTRR455 pKa = 11.84GTSNDD460 pKa = 3.26VAGVALGSSVVINEE474 pKa = 3.82IDD476 pKa = 3.65TVPTSGNDD484 pKa = 3.19HH485 pKa = 6.98LEE487 pKa = 4.38LYY489 pKa = 10.73NPTTTPITVTGWWVSDD505 pKa = 3.61GNDD508 pKa = 3.12WAVITSNIVIPPGGFALLEE527 pKa = 4.45EE528 pKa = 4.52NVDD531 pKa = 3.47WQPEE535 pKa = 4.09DD536 pKa = 3.67FGVDD540 pKa = 4.24FGSTDD545 pKa = 3.12VAYY548 pKa = 10.76LFDD551 pKa = 5.21DD552 pKa = 3.57SFVRR556 pKa = 11.84IDD558 pKa = 3.4QIGWNGEE565 pKa = 3.97AEE567 pKa = 4.45DD568 pKa = 3.84EE569 pKa = 4.51CFARR573 pKa = 11.84VPDD576 pKa = 4.06GAGPNDD582 pKa = 4.26GYY584 pKa = 11.52DD585 pKa = 3.22WLSSGGGSTWLDD597 pKa = 3.38QTCTLGSTNVLSTPGAALAVSKK619 pKa = 9.62ATRR622 pKa = 11.84GLAVIGEE629 pKa = 4.33AHH631 pKa = 6.6TFTITLTTGANDD643 pKa = 3.46APNVVLTDD651 pKa = 3.69TLPVSTTYY659 pKa = 10.9VADD662 pKa = 3.75NSGVTPTMAAPGVYY676 pKa = 8.63VWNFGTVPMSTTITFNLTVTIDD698 pKa = 3.48ANVAAGTVLTNTVEE712 pKa = 4.34VASDD716 pKa = 3.73LAGDD720 pKa = 4.16DD721 pKa = 3.92PADD724 pKa = 3.72NVATATTTAWPLVTIQQIQQVTDD747 pKa = 3.67PATSDD752 pKa = 3.16ASPLVGQSVWVEE764 pKa = 4.05GIVTSEE770 pKa = 4.56PGDD773 pKa = 3.17IDD775 pKa = 3.74TPSRR779 pKa = 11.84IMVIQDD785 pKa = 3.25AAGGPWSGLVLFRR798 pKa = 11.84SAGFDD803 pKa = 3.12PTQFPVGTKK812 pKa = 8.94VRR814 pKa = 11.84AFGTVTEE821 pKa = 4.54YY822 pKa = 11.45YY823 pKa = 10.83GLTEE827 pKa = 4.26LTTTLAEE834 pKa = 4.27PMGTATPPAPQVLNTGAFNDD854 pKa = 3.62ADD856 pKa = 4.16PAVSEE861 pKa = 3.99QWEE864 pKa = 4.67GVLVEE869 pKa = 4.46FQNATVTDD877 pKa = 3.86ADD879 pKa = 4.14LGFGEE884 pKa = 4.38WQFDD888 pKa = 3.96DD889 pKa = 4.66GSGPTRR895 pKa = 11.84ADD897 pKa = 3.45DD898 pKa = 4.84LGEE901 pKa = 4.27KK902 pKa = 10.61DD903 pKa = 4.53GDD905 pKa = 3.84LTYY908 pKa = 11.12LPTLGDD914 pKa = 3.31TYY916 pKa = 11.4GYY918 pKa = 10.12IRR920 pKa = 11.84GIAWYY925 pKa = 10.62SFGNYY930 pKa = 9.14KK931 pKa = 10.47LEE933 pKa = 4.26PRR935 pKa = 11.84SDD937 pKa = 3.35DD938 pKa = 4.22DD939 pKa = 3.44IRR941 pKa = 11.84VVQTTPIVVKK951 pKa = 9.64QAPLNVAPNSVFTYY965 pKa = 9.79TITITNEE972 pKa = 3.23LGYY975 pKa = 10.61DD976 pKa = 3.41LTGVVITDD984 pKa = 5.02RR985 pKa = 11.84IPTANAALATVLDD998 pKa = 4.75GGTLSNGVISWNVGTVADD1016 pKa = 4.06MSSVSVRR1023 pKa = 11.84FVMTATGAMGSVITNDD1039 pKa = 3.01AYY1041 pKa = 10.82GVVAANFITPTLGAPVLTTIGDD1063 pKa = 3.57YY1064 pKa = 10.75TPIYY1068 pKa = 9.07TIQGNDD1074 pKa = 3.17FLSPFRR1080 pKa = 11.84GMPVKK1085 pKa = 10.04TRR1087 pKa = 11.84GVVVGFFEE1095 pKa = 4.85GNYY1098 pKa = 9.23PGSGSFNGFFIQDD1111 pKa = 3.5EE1112 pKa = 4.55TGDD1115 pKa = 4.09GDD1117 pKa = 3.99PATSDD1122 pKa = 3.78GIFVHH1127 pKa = 6.48VGTDD1131 pKa = 3.52TVSPTLKK1138 pKa = 10.17IGDD1141 pKa = 4.03VVTVTGTVEE1150 pKa = 5.3EE1151 pKa = 4.66FIEE1154 pKa = 4.36WDD1156 pKa = 4.03DD1157 pKa = 3.91NACPDD1162 pKa = 4.47TEE1164 pKa = 5.42CITQIRR1170 pKa = 11.84TSMANVQVAGVGSVMPTVLTPVGDD1194 pKa = 4.12PVASEE1199 pKa = 5.03AYY1201 pKa = 8.26WEE1203 pKa = 4.32SLEE1206 pKa = 4.17GMLVTAPNTQTVTGPTSFGAIYY1228 pKa = 9.05VIPGSEE1234 pKa = 3.96GVEE1237 pKa = 3.38RR1238 pKa = 11.84ALRR1241 pKa = 11.84NTPQEE1246 pKa = 3.91GMPVGVRR1253 pKa = 11.84HH1254 pKa = 5.21YY1255 pKa = 10.75EE1256 pKa = 3.7RR1257 pKa = 11.84YY1258 pKa = 10.73GNINGADD1265 pKa = 3.74APSLIVGSVVTDD1277 pKa = 3.09VAGPLGFSYY1286 pKa = 10.96GDD1288 pKa = 3.39YY1289 pKa = 10.44MVITQPGKK1297 pKa = 7.86PWQVVSAKK1305 pKa = 9.53PLPDD1309 pKa = 4.15TPPSWSPPTTEE1320 pKa = 4.16TFTAATFNTLNFDD1333 pKa = 3.92AADD1336 pKa = 3.91PAVKK1340 pKa = 8.09MTKK1343 pKa = 9.75VVSTVLQLGGPTFLALEE1360 pKa = 5.06EE1361 pKa = 4.51IDD1363 pKa = 3.86STTVITDD1370 pKa = 4.71LINNLAAQGIPYY1382 pKa = 9.73AYY1384 pKa = 9.84AASHH1388 pKa = 7.06PDD1390 pKa = 3.38VGGHH1394 pKa = 5.85GVALLWRR1401 pKa = 11.84TDD1403 pKa = 3.39QVTNAVWSTQYY1414 pKa = 10.24QACSPNGSTSAPTYY1428 pKa = 10.68DD1429 pKa = 4.41PYY1431 pKa = 11.58CDD1433 pKa = 4.02AVPGEE1438 pKa = 4.29YY1439 pKa = 9.8PLFSRR1444 pKa = 11.84RR1445 pKa = 11.84PVVLTATVTLNSTPMDD1461 pKa = 3.46VVVIANHH1468 pKa = 6.26FKK1470 pKa = 10.57SKK1472 pKa = 10.6RR1473 pKa = 11.84GGAEE1477 pKa = 3.23SDD1479 pKa = 3.19ARR1481 pKa = 11.84RR1482 pKa = 11.84LEE1484 pKa = 3.79QGQFIGNLAQQLYY1497 pKa = 10.19NNGSRR1502 pKa = 11.84HH1503 pKa = 5.66IIIMGDD1509 pKa = 3.27LNDD1512 pKa = 5.07FEE1514 pKa = 6.23DD1515 pKa = 4.57SPPLQAIYY1523 pKa = 10.57ASGVITSTWYY1533 pKa = 10.07QLPPNEE1539 pKa = 3.99RR1540 pKa = 11.84YY1541 pKa = 9.83SYY1543 pKa = 10.27IFEE1546 pKa = 4.36GVSQVLDD1553 pKa = 3.33HH1554 pKa = 6.61VLISNDD1560 pKa = 3.57LLLALKK1566 pKa = 10.26GVSPLHH1572 pKa = 6.58LNADD1576 pKa = 4.22YY1577 pKa = 10.25PFSPYY1582 pKa = 10.96ADD1584 pKa = 4.12LPVVWQASDD1593 pKa = 3.6HH1594 pKa = 6.79DD1595 pKa = 4.34PVVATFGAIRR1605 pKa = 11.84RR1606 pKa = 11.84IYY1608 pKa = 9.99MPLLFKK1614 pKa = 10.71NAPP1617 pKa = 3.39
MM1 pKa = 7.25VLLLTFSLLVVFIFVPALWVTRR23 pKa = 11.84TTVAHH28 pKa = 6.96AAPEE32 pKa = 4.5GGALIVINEE41 pKa = 4.2FSVKK45 pKa = 9.23STDD48 pKa = 2.92WVEE51 pKa = 4.37LYY53 pKa = 8.48NTTAVTQSVAGWYY66 pKa = 10.45VSDD69 pKa = 4.34TGCGSPTTTMTGTIAPNDD87 pKa = 3.56FRR89 pKa = 11.84VLTVSDD95 pKa = 3.31AGGNFNFSSTAEE107 pKa = 4.22VISLCDD113 pKa = 3.4NAHH116 pKa = 6.01NEE118 pKa = 4.1VDD120 pKa = 3.52RR121 pKa = 11.84VAYY124 pKa = 10.0GIDD127 pKa = 3.31GGAPVPPSNGAFIYY141 pKa = 9.4STARR145 pKa = 11.84VADD148 pKa = 4.24GQDD151 pKa = 3.35TDD153 pKa = 4.99NDD155 pKa = 3.98AADD158 pKa = 3.83WNLDD162 pKa = 3.52STPTPGTPNDD172 pKa = 3.88APGTALGSSLLINEE186 pKa = 3.95IDD188 pKa = 3.86VYY190 pKa = 10.7PDD192 pKa = 3.18SGNDD196 pKa = 3.18IIEE199 pKa = 4.9LYY201 pKa = 10.96NPTTTPITITGWWVSDD217 pKa = 3.41GDD219 pKa = 3.75EE220 pKa = 4.21VAQVVGTVVVEE231 pKa = 4.31PGSVVALEE239 pKa = 4.33VGVDD243 pKa = 3.86LQDD246 pKa = 3.85EE247 pKa = 4.89GTTGLNFTSSDD258 pKa = 3.18VAYY261 pKa = 10.62LFDD264 pKa = 4.16EE265 pKa = 4.0NRR267 pKa = 11.84VRR269 pKa = 11.84VDD271 pKa = 3.09QLGWNGEE278 pKa = 4.11FEE280 pKa = 4.5NGCFARR286 pKa = 11.84VPDD289 pKa = 4.15GAGPNDD295 pKa = 4.26GYY297 pKa = 11.52DD298 pKa = 3.22WLSSGGGATLLDD310 pKa = 4.42LPCSLGAINGGPVVINEE327 pKa = 4.4FAPKK331 pKa = 7.81GTEE334 pKa = 3.53WVEE337 pKa = 3.83LYY339 pKa = 10.57NRR341 pKa = 11.84SPLTQTLTGWYY352 pKa = 8.32LTDD355 pKa = 3.8TACTGATSLITDD367 pKa = 4.47TTPIAPGGFFTVTNGAPGDD386 pKa = 3.87NFALSNNGDD395 pKa = 3.79TLLLCMADD403 pKa = 3.29NTLADD408 pKa = 3.51SVAYY412 pKa = 9.72GNSGGAPLASSTGAGDD428 pKa = 3.55ASIARR433 pKa = 11.84VSDD436 pKa = 3.76GVRR439 pKa = 11.84TGDD442 pKa = 5.69DD443 pKa = 3.33AADD446 pKa = 3.59WNVDD450 pKa = 3.28KK451 pKa = 10.93TPTRR455 pKa = 11.84GTSNDD460 pKa = 3.26VAGVALGSSVVINEE474 pKa = 3.82IDD476 pKa = 3.65TVPTSGNDD484 pKa = 3.19HH485 pKa = 6.98LEE487 pKa = 4.38LYY489 pKa = 10.73NPTTTPITVTGWWVSDD505 pKa = 3.61GNDD508 pKa = 3.12WAVITSNIVIPPGGFALLEE527 pKa = 4.45EE528 pKa = 4.52NVDD531 pKa = 3.47WQPEE535 pKa = 4.09DD536 pKa = 3.67FGVDD540 pKa = 4.24FGSTDD545 pKa = 3.12VAYY548 pKa = 10.76LFDD551 pKa = 5.21DD552 pKa = 3.57SFVRR556 pKa = 11.84IDD558 pKa = 3.4QIGWNGEE565 pKa = 3.97AEE567 pKa = 4.45DD568 pKa = 3.84EE569 pKa = 4.51CFARR573 pKa = 11.84VPDD576 pKa = 4.06GAGPNDD582 pKa = 4.26GYY584 pKa = 11.52DD585 pKa = 3.22WLSSGGGSTWLDD597 pKa = 3.38QTCTLGSTNVLSTPGAALAVSKK619 pKa = 9.62ATRR622 pKa = 11.84GLAVIGEE629 pKa = 4.33AHH631 pKa = 6.6TFTITLTTGANDD643 pKa = 3.46APNVVLTDD651 pKa = 3.69TLPVSTTYY659 pKa = 10.9VADD662 pKa = 3.75NSGVTPTMAAPGVYY676 pKa = 8.63VWNFGTVPMSTTITFNLTVTIDD698 pKa = 3.48ANVAAGTVLTNTVEE712 pKa = 4.34VASDD716 pKa = 3.73LAGDD720 pKa = 4.16DD721 pKa = 3.92PADD724 pKa = 3.72NVATATTTAWPLVTIQQIQQVTDD747 pKa = 3.67PATSDD752 pKa = 3.16ASPLVGQSVWVEE764 pKa = 4.05GIVTSEE770 pKa = 4.56PGDD773 pKa = 3.17IDD775 pKa = 3.74TPSRR779 pKa = 11.84IMVIQDD785 pKa = 3.25AAGGPWSGLVLFRR798 pKa = 11.84SAGFDD803 pKa = 3.12PTQFPVGTKK812 pKa = 8.94VRR814 pKa = 11.84AFGTVTEE821 pKa = 4.54YY822 pKa = 11.45YY823 pKa = 10.83GLTEE827 pKa = 4.26LTTTLAEE834 pKa = 4.27PMGTATPPAPQVLNTGAFNDD854 pKa = 3.62ADD856 pKa = 4.16PAVSEE861 pKa = 3.99QWEE864 pKa = 4.67GVLVEE869 pKa = 4.46FQNATVTDD877 pKa = 3.86ADD879 pKa = 4.14LGFGEE884 pKa = 4.38WQFDD888 pKa = 3.96DD889 pKa = 4.66GSGPTRR895 pKa = 11.84ADD897 pKa = 3.45DD898 pKa = 4.84LGEE901 pKa = 4.27KK902 pKa = 10.61DD903 pKa = 4.53GDD905 pKa = 3.84LTYY908 pKa = 11.12LPTLGDD914 pKa = 3.31TYY916 pKa = 11.4GYY918 pKa = 10.12IRR920 pKa = 11.84GIAWYY925 pKa = 10.62SFGNYY930 pKa = 9.14KK931 pKa = 10.47LEE933 pKa = 4.26PRR935 pKa = 11.84SDD937 pKa = 3.35DD938 pKa = 4.22DD939 pKa = 3.44IRR941 pKa = 11.84VVQTTPIVVKK951 pKa = 9.64QAPLNVAPNSVFTYY965 pKa = 9.79TITITNEE972 pKa = 3.23LGYY975 pKa = 10.61DD976 pKa = 3.41LTGVVITDD984 pKa = 5.02RR985 pKa = 11.84IPTANAALATVLDD998 pKa = 4.75GGTLSNGVISWNVGTVADD1016 pKa = 4.06MSSVSVRR1023 pKa = 11.84FVMTATGAMGSVITNDD1039 pKa = 3.01AYY1041 pKa = 10.82GVVAANFITPTLGAPVLTTIGDD1063 pKa = 3.57YY1064 pKa = 10.75TPIYY1068 pKa = 9.07TIQGNDD1074 pKa = 3.17FLSPFRR1080 pKa = 11.84GMPVKK1085 pKa = 10.04TRR1087 pKa = 11.84GVVVGFFEE1095 pKa = 4.85GNYY1098 pKa = 9.23PGSGSFNGFFIQDD1111 pKa = 3.5EE1112 pKa = 4.55TGDD1115 pKa = 4.09GDD1117 pKa = 3.99PATSDD1122 pKa = 3.78GIFVHH1127 pKa = 6.48VGTDD1131 pKa = 3.52TVSPTLKK1138 pKa = 10.17IGDD1141 pKa = 4.03VVTVTGTVEE1150 pKa = 5.3EE1151 pKa = 4.66FIEE1154 pKa = 4.36WDD1156 pKa = 4.03DD1157 pKa = 3.91NACPDD1162 pKa = 4.47TEE1164 pKa = 5.42CITQIRR1170 pKa = 11.84TSMANVQVAGVGSVMPTVLTPVGDD1194 pKa = 4.12PVASEE1199 pKa = 5.03AYY1201 pKa = 8.26WEE1203 pKa = 4.32SLEE1206 pKa = 4.17GMLVTAPNTQTVTGPTSFGAIYY1228 pKa = 9.05VIPGSEE1234 pKa = 3.96GVEE1237 pKa = 3.38RR1238 pKa = 11.84ALRR1241 pKa = 11.84NTPQEE1246 pKa = 3.91GMPVGVRR1253 pKa = 11.84HH1254 pKa = 5.21YY1255 pKa = 10.75EE1256 pKa = 3.7RR1257 pKa = 11.84YY1258 pKa = 10.73GNINGADD1265 pKa = 3.74APSLIVGSVVTDD1277 pKa = 3.09VAGPLGFSYY1286 pKa = 10.96GDD1288 pKa = 3.39YY1289 pKa = 10.44MVITQPGKK1297 pKa = 7.86PWQVVSAKK1305 pKa = 9.53PLPDD1309 pKa = 4.15TPPSWSPPTTEE1320 pKa = 4.16TFTAATFNTLNFDD1333 pKa = 3.92AADD1336 pKa = 3.91PAVKK1340 pKa = 8.09MTKK1343 pKa = 9.75VVSTVLQLGGPTFLALEE1360 pKa = 5.06EE1361 pKa = 4.51IDD1363 pKa = 3.86STTVITDD1370 pKa = 4.71LINNLAAQGIPYY1382 pKa = 9.73AYY1384 pKa = 9.84AASHH1388 pKa = 7.06PDD1390 pKa = 3.38VGGHH1394 pKa = 5.85GVALLWRR1401 pKa = 11.84TDD1403 pKa = 3.39QVTNAVWSTQYY1414 pKa = 10.24QACSPNGSTSAPTYY1428 pKa = 10.68DD1429 pKa = 4.41PYY1431 pKa = 11.58CDD1433 pKa = 4.02AVPGEE1438 pKa = 4.29YY1439 pKa = 9.8PLFSRR1444 pKa = 11.84RR1445 pKa = 11.84PVVLTATVTLNSTPMDD1461 pKa = 3.46VVVIANHH1468 pKa = 6.26FKK1470 pKa = 10.57SKK1472 pKa = 10.6RR1473 pKa = 11.84GGAEE1477 pKa = 3.23SDD1479 pKa = 3.19ARR1481 pKa = 11.84RR1482 pKa = 11.84LEE1484 pKa = 3.79QGQFIGNLAQQLYY1497 pKa = 10.19NNGSRR1502 pKa = 11.84HH1503 pKa = 5.66IIIMGDD1509 pKa = 3.27LNDD1512 pKa = 5.07FEE1514 pKa = 6.23DD1515 pKa = 4.57SPPLQAIYY1523 pKa = 10.57ASGVITSTWYY1533 pKa = 10.07QLPPNEE1539 pKa = 3.99RR1540 pKa = 11.84YY1541 pKa = 9.83SYY1543 pKa = 10.27IFEE1546 pKa = 4.36GVSQVLDD1553 pKa = 3.33HH1554 pKa = 6.61VLISNDD1560 pKa = 3.57LLLALKK1566 pKa = 10.26GVSPLHH1572 pKa = 6.58LNADD1576 pKa = 4.22YY1577 pKa = 10.25PFSPYY1582 pKa = 10.96ADD1584 pKa = 4.12LPVVWQASDD1593 pKa = 3.6HH1594 pKa = 6.79DD1595 pKa = 4.34PVVATFGAIRR1605 pKa = 11.84RR1606 pKa = 11.84IYY1608 pKa = 9.99MPLLFKK1614 pKa = 10.71NAPP1617 pKa = 3.39
Molecular weight: 170.32 kDa
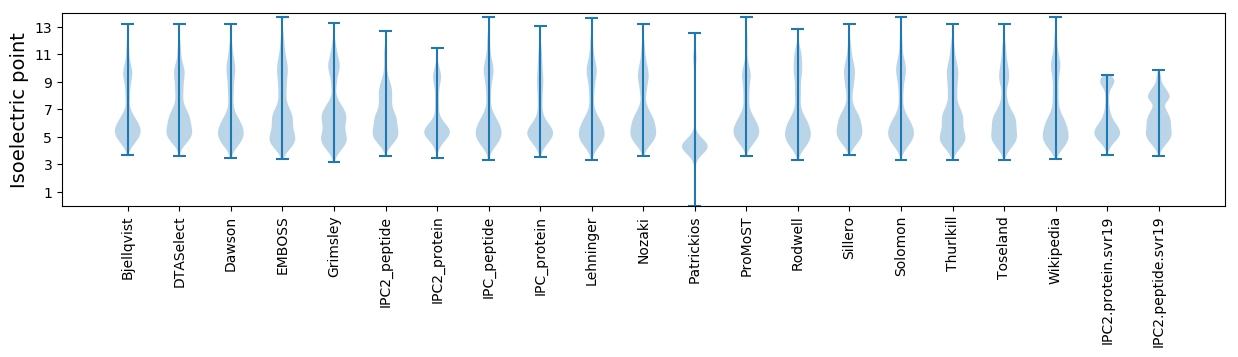
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M8K942|A0A0M8K942_9CHLR Uncharacterized protein OS=Ardenticatena maritima OX=872965 GN=ARMA_2756 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 8.95RR4 pKa = 11.84TLIRR8 pKa = 11.84KK9 pKa = 7.74RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84MRR15 pKa = 11.84VHH17 pKa = 6.88GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TPGGRR29 pKa = 11.84RR30 pKa = 11.84VLKK33 pKa = 9.72NRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.52GRR40 pKa = 11.84HH41 pKa = 3.96QLTVKK46 pKa = 10.36IPRR49 pKa = 11.84RR50 pKa = 11.84WFF52 pKa = 3.38
MM1 pKa = 8.0PKK3 pKa = 8.95RR4 pKa = 11.84TLIRR8 pKa = 11.84KK9 pKa = 7.74RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84MRR15 pKa = 11.84VHH17 pKa = 6.88GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TPGGRR29 pKa = 11.84RR30 pKa = 11.84VLKK33 pKa = 9.72NRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.52GRR40 pKa = 11.84HH41 pKa = 3.96QLTVKK46 pKa = 10.36IPRR49 pKa = 11.84RR50 pKa = 11.84WFF52 pKa = 3.38
Molecular weight: 6.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1012731 |
37 |
2171 |
322.1 |
35.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.093 ± 0.054 | 0.694 ± 0.013 |
5.138 ± 0.032 | 6.63 ± 0.059 |
3.763 ± 0.03 | 7.284 ± 0.046 |
2.564 ± 0.024 | 5.113 ± 0.033 |
2.368 ± 0.03 | 10.98 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.224 ± 0.021 | 2.634 ± 0.028 |
5.808 ± 0.035 | 3.658 ± 0.026 |
7.512 ± 0.047 | 4.08 ± 0.033 |
5.878 ± 0.034 | 7.962 ± 0.038 |
1.806 ± 0.025 | 2.764 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
