
Agrococcus casei LMG 22410
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Agrococcus; Agrococcus casei
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
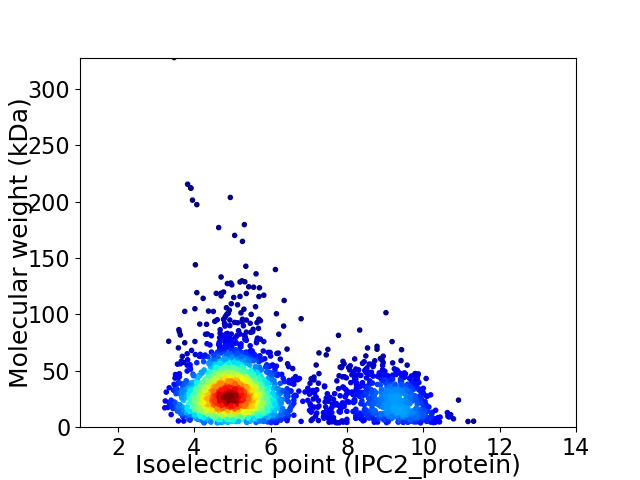
Virtual 2D-PAGE plot for 2698 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1R4GJN6|A0A1R4GJN6_9MICO Chromosome (Plasmid) partitioning protein ParA OS=Agrococcus casei LMG 22410 OX=1255656 GN=CZ674_12595 PE=4 SV=1
MM1 pKa = 7.0IHH3 pKa = 6.46VRR5 pKa = 11.84TLTKK9 pKa = 10.56KK10 pKa = 10.48LGIVALSATVAASGLTAITPAPAAQAAPSTGLVISEE46 pKa = 4.51AYY48 pKa = 10.29GGGGNGGAKK57 pKa = 10.12FNQDD61 pKa = 3.72FIEE64 pKa = 4.97LYY66 pKa = 9.74NFSAAEE72 pKa = 3.89VSLDD76 pKa = 3.26GVTVSYY82 pKa = 10.85YY83 pKa = 10.69SSSGNLGASTEE94 pKa = 4.49LEE96 pKa = 4.25GSLQPGEE103 pKa = 4.55HH104 pKa = 6.29YY105 pKa = 10.87LVGAAYY111 pKa = 10.04GSNRR115 pKa = 11.84DD116 pKa = 3.61LPSFEE121 pKa = 5.13SDD123 pKa = 2.83ATFNAAMSGSKK134 pKa = 10.69GSVEE138 pKa = 3.68IAAGDD143 pKa = 3.83TVIDD147 pKa = 3.86LVGWGDD153 pKa = 3.19AALFEE158 pKa = 4.73GAPAHH163 pKa = 5.57GTSNSVSVARR173 pKa = 11.84VDD175 pKa = 3.81PASDD179 pKa = 3.39TDD181 pKa = 4.06DD182 pKa = 3.91NSVDD186 pKa = 3.96FATGEE191 pKa = 4.01PTPQGLAEE199 pKa = 5.64DD200 pKa = 4.47GTEE203 pKa = 4.3EE204 pKa = 4.71PPVDD208 pKa = 4.05PPADD212 pKa = 3.87PEE214 pKa = 4.14TATISEE220 pKa = 4.25IQGEE224 pKa = 4.49GAEE227 pKa = 4.62TPFMDD232 pKa = 4.94QPVITQGIVTAVYY245 pKa = 9.01TGQGSKK251 pKa = 10.75NGFNIQEE258 pKa = 4.37PGVVDD263 pKa = 4.04IASHH267 pKa = 5.92AASTGVFVYY276 pKa = 10.34GSSHH280 pKa = 6.59AEE282 pKa = 3.78SVQIGDD288 pKa = 3.71SVEE291 pKa = 3.97VHH293 pKa = 5.61GTATEE298 pKa = 4.47YY299 pKa = 11.53YY300 pKa = 10.25DD301 pKa = 3.45VTQIEE306 pKa = 4.42ASSVTALGEE315 pKa = 4.34SLGEE319 pKa = 3.89ATAVEE324 pKa = 4.24LAEE327 pKa = 4.18WPTEE331 pKa = 4.01EE332 pKa = 4.18EE333 pKa = 3.84VRR335 pKa = 11.84EE336 pKa = 4.13LFEE339 pKa = 5.34GMLLAPQGTYY349 pKa = 8.7TVSNNYY355 pKa = 10.11ALNQYY360 pKa = 10.54GEE362 pKa = 4.31VGLAFGDD369 pKa = 4.0SALVQPTEE377 pKa = 4.02VGAPGSDD384 pKa = 2.91AAYY387 pKa = 10.22AQAEE391 pKa = 4.3ANAAASVLLDD401 pKa = 5.59DD402 pKa = 4.8GQTWDD407 pKa = 4.01YY408 pKa = 10.1MSNDD412 pKa = 3.52AAKK415 pKa = 10.39DD416 pKa = 3.55SPLPYY421 pKa = 9.35LTLEE425 pKa = 4.22TPVTIGAVAAFTDD438 pKa = 4.89GVVLDD443 pKa = 4.59YY444 pKa = 11.76SHH446 pKa = 6.34GTWRR450 pKa = 11.84FQPTQPVHH458 pKa = 5.05GTEE461 pKa = 3.93NAPVAFSDD469 pKa = 3.74VRR471 pKa = 11.84EE472 pKa = 4.04QAPEE476 pKa = 3.95DD477 pKa = 3.51VGGDD481 pKa = 3.23ITIGTFNVLNYY492 pKa = 7.4FTTLGDD498 pKa = 3.68TEE500 pKa = 4.86AGCEE504 pKa = 3.93AYY506 pKa = 9.79EE507 pKa = 4.9DD508 pKa = 4.4RR509 pKa = 11.84DD510 pKa = 3.96GNPIATDD517 pKa = 3.39YY518 pKa = 7.54CTPRR522 pKa = 11.84GAYY525 pKa = 9.6DD526 pKa = 3.24AEE528 pKa = 4.19NFEE531 pKa = 4.28RR532 pKa = 11.84QQAKK536 pKa = 9.68IVTAINTLDD545 pKa = 3.49ASVVALEE552 pKa = 4.69EE553 pKa = 4.57IEE555 pKa = 4.83DD556 pKa = 3.9SSDD559 pKa = 3.27FGNDD563 pKa = 2.42RR564 pKa = 11.84DD565 pKa = 3.7ASIEE569 pKa = 4.07HH570 pKa = 6.55LVGEE574 pKa = 4.69LNAAADD580 pKa = 3.94EE581 pKa = 4.47DD582 pKa = 3.65RR583 pKa = 11.84WAFVPSPEE591 pKa = 4.52WIPATGDD598 pKa = 3.2DD599 pKa = 4.56VIRR602 pKa = 11.84TGYY605 pKa = 9.32IYY607 pKa = 10.77KK608 pKa = 9.67PAEE611 pKa = 3.58VSYY614 pKa = 11.08IEE616 pKa = 4.84GSAEE620 pKa = 4.09LLDD623 pKa = 4.71DD624 pKa = 4.67AAFEE628 pKa = 4.07NARR631 pKa = 11.84APFAALFAPADD642 pKa = 3.72EE643 pKa = 4.62ALSGNEE649 pKa = 3.51FLVIANHH656 pKa = 6.31FKK658 pKa = 11.12SKK660 pKa = 10.52GGSGDD665 pKa = 3.27GDD667 pKa = 3.48NANRR671 pKa = 11.84DD672 pKa = 3.92DD673 pKa = 4.38EE674 pKa = 5.38LGPAHH679 pKa = 7.26AVGGWNGDD687 pKa = 3.17RR688 pKa = 11.84VRR690 pKa = 11.84QAEE693 pKa = 4.26AVVEE697 pKa = 4.38FADD700 pKa = 4.5TLEE703 pKa = 4.26QQHH706 pKa = 5.73GTDD709 pKa = 3.86LVFITGDD716 pKa = 3.54LNSYY720 pKa = 9.17AQEE723 pKa = 4.48DD724 pKa = 4.6PVTTILNAGYY734 pKa = 8.77TNLTAGGPEE743 pKa = 3.84YY744 pKa = 10.75SYY746 pKa = 11.74AFGGMVGSLDD756 pKa = 3.26HH757 pKa = 6.64VLASEE762 pKa = 4.18AATALVTGADD772 pKa = 3.28LWTINAYY779 pKa = 10.06EE780 pKa = 4.14PVALEE785 pKa = 4.07YY786 pKa = 10.77SRR788 pKa = 11.84HH789 pKa = 5.51NYY791 pKa = 9.11NVAQLYY797 pKa = 10.36AADD800 pKa = 3.83QFRR803 pKa = 11.84SSDD806 pKa = 3.64HH807 pKa = 6.34NPALVGVSFAEE818 pKa = 4.7DD819 pKa = 3.5DD820 pKa = 5.05DD821 pKa = 5.18IEE823 pKa = 5.96ADD825 pKa = 3.68EE826 pKa = 4.6PLLVIEE832 pKa = 4.37QEE834 pKa = 4.34QYY836 pKa = 9.58TQTEE840 pKa = 4.62SIDD843 pKa = 3.79GVSYY847 pKa = 10.73AATGLRR853 pKa = 11.84EE854 pKa = 3.97GTAVVEE860 pKa = 4.86LIDD863 pKa = 3.97AGGSVSVIEE872 pKa = 4.87GAAQTEE878 pKa = 4.29AGALAGEE885 pKa = 4.37VSYY888 pKa = 8.88EE889 pKa = 4.07TEE891 pKa = 3.63AGEE894 pKa = 4.3RR895 pKa = 11.84LRR897 pKa = 11.84MPVGTYY903 pKa = 7.07TLRR906 pKa = 11.84VTQEE910 pKa = 3.71GSGGAEE916 pKa = 3.92SIVLEE921 pKa = 4.09AAFEE925 pKa = 4.32VVADD929 pKa = 3.95EE930 pKa = 4.77TGDD933 pKa = 4.69DD934 pKa = 3.84GTGDD938 pKa = 4.0DD939 pKa = 4.69GSFDD943 pKa = 3.92GDD945 pKa = 3.25EE946 pKa = 5.1DD947 pKa = 4.17GQPDD951 pKa = 3.84LPEE954 pKa = 4.15TGFDD958 pKa = 3.15GGLAPFVLALALMLGGTGFVLLRR981 pKa = 11.84RR982 pKa = 11.84RR983 pKa = 11.84NAA985 pKa = 3.24
MM1 pKa = 7.0IHH3 pKa = 6.46VRR5 pKa = 11.84TLTKK9 pKa = 10.56KK10 pKa = 10.48LGIVALSATVAASGLTAITPAPAAQAAPSTGLVISEE46 pKa = 4.51AYY48 pKa = 10.29GGGGNGGAKK57 pKa = 10.12FNQDD61 pKa = 3.72FIEE64 pKa = 4.97LYY66 pKa = 9.74NFSAAEE72 pKa = 3.89VSLDD76 pKa = 3.26GVTVSYY82 pKa = 10.85YY83 pKa = 10.69SSSGNLGASTEE94 pKa = 4.49LEE96 pKa = 4.25GSLQPGEE103 pKa = 4.55HH104 pKa = 6.29YY105 pKa = 10.87LVGAAYY111 pKa = 10.04GSNRR115 pKa = 11.84DD116 pKa = 3.61LPSFEE121 pKa = 5.13SDD123 pKa = 2.83ATFNAAMSGSKK134 pKa = 10.69GSVEE138 pKa = 3.68IAAGDD143 pKa = 3.83TVIDD147 pKa = 3.86LVGWGDD153 pKa = 3.19AALFEE158 pKa = 4.73GAPAHH163 pKa = 5.57GTSNSVSVARR173 pKa = 11.84VDD175 pKa = 3.81PASDD179 pKa = 3.39TDD181 pKa = 4.06DD182 pKa = 3.91NSVDD186 pKa = 3.96FATGEE191 pKa = 4.01PTPQGLAEE199 pKa = 5.64DD200 pKa = 4.47GTEE203 pKa = 4.3EE204 pKa = 4.71PPVDD208 pKa = 4.05PPADD212 pKa = 3.87PEE214 pKa = 4.14TATISEE220 pKa = 4.25IQGEE224 pKa = 4.49GAEE227 pKa = 4.62TPFMDD232 pKa = 4.94QPVITQGIVTAVYY245 pKa = 9.01TGQGSKK251 pKa = 10.75NGFNIQEE258 pKa = 4.37PGVVDD263 pKa = 4.04IASHH267 pKa = 5.92AASTGVFVYY276 pKa = 10.34GSSHH280 pKa = 6.59AEE282 pKa = 3.78SVQIGDD288 pKa = 3.71SVEE291 pKa = 3.97VHH293 pKa = 5.61GTATEE298 pKa = 4.47YY299 pKa = 11.53YY300 pKa = 10.25DD301 pKa = 3.45VTQIEE306 pKa = 4.42ASSVTALGEE315 pKa = 4.34SLGEE319 pKa = 3.89ATAVEE324 pKa = 4.24LAEE327 pKa = 4.18WPTEE331 pKa = 4.01EE332 pKa = 4.18EE333 pKa = 3.84VRR335 pKa = 11.84EE336 pKa = 4.13LFEE339 pKa = 5.34GMLLAPQGTYY349 pKa = 8.7TVSNNYY355 pKa = 10.11ALNQYY360 pKa = 10.54GEE362 pKa = 4.31VGLAFGDD369 pKa = 4.0SALVQPTEE377 pKa = 4.02VGAPGSDD384 pKa = 2.91AAYY387 pKa = 10.22AQAEE391 pKa = 4.3ANAAASVLLDD401 pKa = 5.59DD402 pKa = 4.8GQTWDD407 pKa = 4.01YY408 pKa = 10.1MSNDD412 pKa = 3.52AAKK415 pKa = 10.39DD416 pKa = 3.55SPLPYY421 pKa = 9.35LTLEE425 pKa = 4.22TPVTIGAVAAFTDD438 pKa = 4.89GVVLDD443 pKa = 4.59YY444 pKa = 11.76SHH446 pKa = 6.34GTWRR450 pKa = 11.84FQPTQPVHH458 pKa = 5.05GTEE461 pKa = 3.93NAPVAFSDD469 pKa = 3.74VRR471 pKa = 11.84EE472 pKa = 4.04QAPEE476 pKa = 3.95DD477 pKa = 3.51VGGDD481 pKa = 3.23ITIGTFNVLNYY492 pKa = 7.4FTTLGDD498 pKa = 3.68TEE500 pKa = 4.86AGCEE504 pKa = 3.93AYY506 pKa = 9.79EE507 pKa = 4.9DD508 pKa = 4.4RR509 pKa = 11.84DD510 pKa = 3.96GNPIATDD517 pKa = 3.39YY518 pKa = 7.54CTPRR522 pKa = 11.84GAYY525 pKa = 9.6DD526 pKa = 3.24AEE528 pKa = 4.19NFEE531 pKa = 4.28RR532 pKa = 11.84QQAKK536 pKa = 9.68IVTAINTLDD545 pKa = 3.49ASVVALEE552 pKa = 4.69EE553 pKa = 4.57IEE555 pKa = 4.83DD556 pKa = 3.9SSDD559 pKa = 3.27FGNDD563 pKa = 2.42RR564 pKa = 11.84DD565 pKa = 3.7ASIEE569 pKa = 4.07HH570 pKa = 6.55LVGEE574 pKa = 4.69LNAAADD580 pKa = 3.94EE581 pKa = 4.47DD582 pKa = 3.65RR583 pKa = 11.84WAFVPSPEE591 pKa = 4.52WIPATGDD598 pKa = 3.2DD599 pKa = 4.56VIRR602 pKa = 11.84TGYY605 pKa = 9.32IYY607 pKa = 10.77KK608 pKa = 9.67PAEE611 pKa = 3.58VSYY614 pKa = 11.08IEE616 pKa = 4.84GSAEE620 pKa = 4.09LLDD623 pKa = 4.71DD624 pKa = 4.67AAFEE628 pKa = 4.07NARR631 pKa = 11.84APFAALFAPADD642 pKa = 3.72EE643 pKa = 4.62ALSGNEE649 pKa = 3.51FLVIANHH656 pKa = 6.31FKK658 pKa = 11.12SKK660 pKa = 10.52GGSGDD665 pKa = 3.27GDD667 pKa = 3.48NANRR671 pKa = 11.84DD672 pKa = 3.92DD673 pKa = 4.38EE674 pKa = 5.38LGPAHH679 pKa = 7.26AVGGWNGDD687 pKa = 3.17RR688 pKa = 11.84VRR690 pKa = 11.84QAEE693 pKa = 4.26AVVEE697 pKa = 4.38FADD700 pKa = 4.5TLEE703 pKa = 4.26QQHH706 pKa = 5.73GTDD709 pKa = 3.86LVFITGDD716 pKa = 3.54LNSYY720 pKa = 9.17AQEE723 pKa = 4.48DD724 pKa = 4.6PVTTILNAGYY734 pKa = 8.77TNLTAGGPEE743 pKa = 3.84YY744 pKa = 10.75SYY746 pKa = 11.74AFGGMVGSLDD756 pKa = 3.26HH757 pKa = 6.64VLASEE762 pKa = 4.18AATALVTGADD772 pKa = 3.28LWTINAYY779 pKa = 10.06EE780 pKa = 4.14PVALEE785 pKa = 4.07YY786 pKa = 10.77SRR788 pKa = 11.84HH789 pKa = 5.51NYY791 pKa = 9.11NVAQLYY797 pKa = 10.36AADD800 pKa = 3.83QFRR803 pKa = 11.84SSDD806 pKa = 3.64HH807 pKa = 6.34NPALVGVSFAEE818 pKa = 4.7DD819 pKa = 3.5DD820 pKa = 5.05DD821 pKa = 5.18IEE823 pKa = 5.96ADD825 pKa = 3.68EE826 pKa = 4.6PLLVIEE832 pKa = 4.37QEE834 pKa = 4.34QYY836 pKa = 9.58TQTEE840 pKa = 4.62SIDD843 pKa = 3.79GVSYY847 pKa = 10.73AATGLRR853 pKa = 11.84EE854 pKa = 3.97GTAVVEE860 pKa = 4.86LIDD863 pKa = 3.97AGGSVSVIEE872 pKa = 4.87GAAQTEE878 pKa = 4.29AGALAGEE885 pKa = 4.37VSYY888 pKa = 8.88EE889 pKa = 4.07TEE891 pKa = 3.63AGEE894 pKa = 4.3RR895 pKa = 11.84LRR897 pKa = 11.84MPVGTYY903 pKa = 7.07TLRR906 pKa = 11.84VTQEE910 pKa = 3.71GSGGAEE916 pKa = 3.92SIVLEE921 pKa = 4.09AAFEE925 pKa = 4.32VVADD929 pKa = 3.95EE930 pKa = 4.77TGDD933 pKa = 4.69DD934 pKa = 3.84GTGDD938 pKa = 4.0DD939 pKa = 4.69GSFDD943 pKa = 3.92GDD945 pKa = 3.25EE946 pKa = 5.1DD947 pKa = 4.17GQPDD951 pKa = 3.84LPEE954 pKa = 4.15TGFDD958 pKa = 3.15GGLAPFVLALALMLGGTGFVLLRR981 pKa = 11.84RR982 pKa = 11.84RR983 pKa = 11.84NAA985 pKa = 3.24
Molecular weight: 102.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1R4ERB9|A0A1R4ERB9_9MICO Uncharacterized protein OS=Agrococcus casei LMG 22410 OX=1255656 GN=CZ674_00375 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.74GRR40 pKa = 11.84SSVSAA45 pKa = 3.76
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.74GRR40 pKa = 11.84SSVSAA45 pKa = 3.76
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
827534 |
37 |
3196 |
306.7 |
33.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.571 ± 0.067 | 0.483 ± 0.011 |
6.297 ± 0.046 | 6.41 ± 0.052 |
3.181 ± 0.034 | 8.588 ± 0.041 |
2.095 ± 0.022 | 4.917 ± 0.035 |
2.469 ± 0.038 | 9.724 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.199 ± 0.023 | 2.198 ± 0.03 |
4.843 ± 0.039 | 3.436 ± 0.027 |
6.705 ± 0.061 | 5.991 ± 0.031 |
6.0 ± 0.042 | 8.545 ± 0.041 |
1.43 ± 0.025 | 1.917 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
