
Collichthys lucidus (Big head croaker) (Sciaena lucida)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei; Eurypterygia;
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
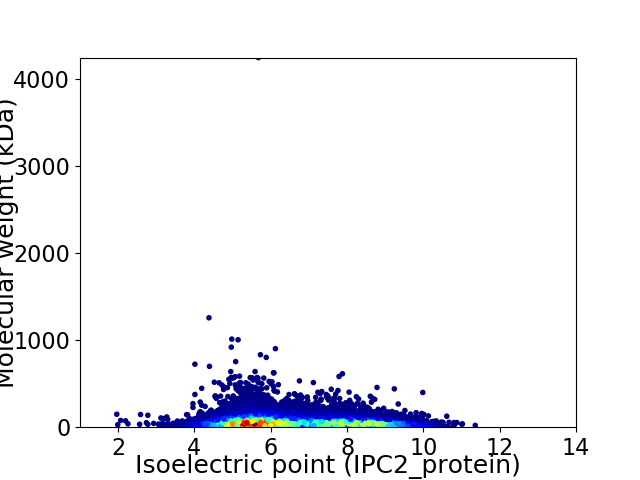
Virtual 2D-PAGE plot for 27208 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
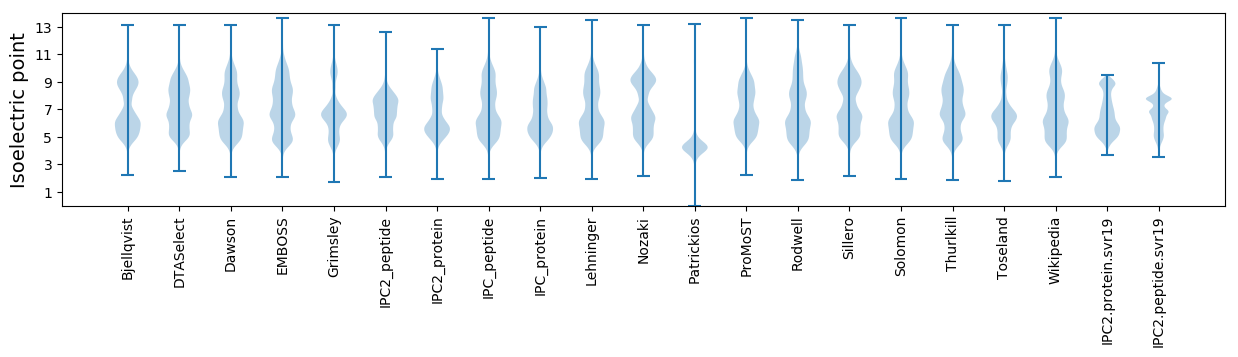
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4U5TVY1|A0A4U5TVY1_COLLU Protein unc-80-like protein OS=Collichthys lucidus OX=240159 GN=D9C73_027713 PE=4 SV=1
MM1 pKa = 7.96IDD3 pKa = 4.76RR4 pKa = 11.84DD5 pKa = 4.14MIDD8 pKa = 4.84CDD10 pKa = 5.55VIDD13 pKa = 5.85CDD15 pKa = 5.73MIDD18 pKa = 4.12CDD20 pKa = 4.44VFDD23 pKa = 5.56RR24 pKa = 11.84DD25 pKa = 3.67VFDD28 pKa = 5.68RR29 pKa = 11.84DD30 pKa = 3.45VFDD33 pKa = 5.68RR34 pKa = 11.84DD35 pKa = 3.45VFDD38 pKa = 6.19RR39 pKa = 11.84DD40 pKa = 4.02VFDD43 pKa = 6.24CDD45 pKa = 5.14VFDD48 pKa = 6.49CDD50 pKa = 5.84VFDD53 pKa = 6.49CDD55 pKa = 5.84VFDD58 pKa = 5.74CDD60 pKa = 4.46VFDD63 pKa = 6.55RR64 pKa = 11.84DD65 pKa = 4.94VFDD68 pKa = 5.6CDD70 pKa = 3.53VFDD73 pKa = 6.55RR74 pKa = 11.84DD75 pKa = 4.94VFDD78 pKa = 5.6CDD80 pKa = 3.53VFDD83 pKa = 6.55RR84 pKa = 11.84DD85 pKa = 4.94VFDD88 pKa = 6.24CDD90 pKa = 5.14VFDD93 pKa = 5.74CDD95 pKa = 4.46VFDD98 pKa = 5.74RR99 pKa = 11.84DD100 pKa = 3.67VFDD103 pKa = 6.19RR104 pKa = 11.84DD105 pKa = 4.02VFDD108 pKa = 5.6CDD110 pKa = 3.53VFDD113 pKa = 6.55RR114 pKa = 11.84DD115 pKa = 4.94VFDD118 pKa = 6.24CDD120 pKa = 5.14VFDD123 pKa = 5.74CDD125 pKa = 4.46VFDD128 pKa = 6.55RR129 pKa = 11.84DD130 pKa = 4.94VFDD133 pKa = 5.6CDD135 pKa = 3.53VFDD138 pKa = 6.55RR139 pKa = 11.84DD140 pKa = 4.94VFDD143 pKa = 5.6CDD145 pKa = 3.53VFDD148 pKa = 6.55RR149 pKa = 11.84DD150 pKa = 4.94VFDD153 pKa = 5.6CDD155 pKa = 3.53VFDD158 pKa = 6.55RR159 pKa = 11.84DD160 pKa = 4.94VFDD163 pKa = 5.6CDD165 pKa = 3.53VFDD168 pKa = 5.74RR169 pKa = 11.84DD170 pKa = 3.67VFDD173 pKa = 5.68RR174 pKa = 11.84DD175 pKa = 3.45VFDD178 pKa = 5.68RR179 pKa = 11.84DD180 pKa = 3.45VFDD183 pKa = 6.19RR184 pKa = 11.84DD185 pKa = 4.02VFDD188 pKa = 6.24CDD190 pKa = 5.14VFDD193 pKa = 5.74CDD195 pKa = 4.46VFDD198 pKa = 6.55RR199 pKa = 11.84DD200 pKa = 4.94VFDD203 pKa = 6.24CDD205 pKa = 5.14VFDD208 pKa = 6.49CDD210 pKa = 5.84VFDD213 pKa = 5.74CDD215 pKa = 4.46VFDD218 pKa = 6.55RR219 pKa = 11.84DD220 pKa = 4.94VFDD223 pKa = 5.6CDD225 pKa = 3.49VFDD228 pKa = 6.49RR229 pKa = 11.84DD230 pKa = 5.38VIDD233 pKa = 5.75CDD235 pKa = 4.82VFDD238 pKa = 5.81CDD240 pKa = 5.13VIDD243 pKa = 5.91RR244 pKa = 11.84DD245 pKa = 4.62VFDD248 pKa = 5.66CDD250 pKa = 4.13VIDD253 pKa = 4.92RR254 pKa = 11.84DD255 pKa = 4.15VIDD258 pKa = 5.57RR259 pKa = 11.84DD260 pKa = 4.0VFDD263 pKa = 6.13CDD265 pKa = 4.55VFDD268 pKa = 5.74CNVIDD273 pKa = 6.05RR274 pKa = 11.84DD275 pKa = 4.14VFDD278 pKa = 5.66CDD280 pKa = 4.27VIDD283 pKa = 4.72RR284 pKa = 11.84DD285 pKa = 3.86VFDD288 pKa = 6.07RR289 pKa = 11.84DD290 pKa = 3.94VFDD293 pKa = 5.55CDD295 pKa = 4.27VIDD298 pKa = 5.91RR299 pKa = 11.84DD300 pKa = 4.79VFDD303 pKa = 6.04CDD305 pKa = 5.14VFDD308 pKa = 6.49CDD310 pKa = 5.84VFDD313 pKa = 6.49CDD315 pKa = 5.84VFDD318 pKa = 6.49CDD320 pKa = 5.84VFDD323 pKa = 6.49CDD325 pKa = 5.84VFDD328 pKa = 5.74CDD330 pKa = 4.46VFDD333 pKa = 5.74RR334 pKa = 11.84DD335 pKa = 3.67VFDD338 pKa = 6.19RR339 pKa = 11.84DD340 pKa = 4.02VFDD343 pKa = 6.24CDD345 pKa = 5.14VFDD348 pKa = 5.74CDD350 pKa = 4.26VFDD353 pKa = 5.78RR354 pKa = 11.84DD355 pKa = 4.18VIDD358 pKa = 4.79RR359 pKa = 11.84DD360 pKa = 3.69VFDD363 pKa = 5.72RR364 pKa = 11.84DD365 pKa = 3.45VFDD368 pKa = 5.68RR369 pKa = 11.84DD370 pKa = 3.45VFDD373 pKa = 5.68RR374 pKa = 11.84DD375 pKa = 3.45VFDD378 pKa = 5.68RR379 pKa = 11.84DD380 pKa = 3.45VFDD383 pKa = 5.68RR384 pKa = 11.84DD385 pKa = 3.45VFDD388 pKa = 6.19RR389 pKa = 11.84DD390 pKa = 4.02VFDD393 pKa = 6.24CDD395 pKa = 5.14VFDD398 pKa = 6.49CDD400 pKa = 5.84VFDD403 pKa = 5.6CDD405 pKa = 5.14VFDD408 pKa = 6.11FMNHH412 pKa = 6.56DD413 pKa = 4.18YY414 pKa = 10.18MNHH417 pKa = 4.38STMPMKK423 pKa = 10.3HH424 pKa = 6.36DD425 pKa = 3.4HH426 pKa = 6.93HH427 pKa = 7.38NDD429 pKa = 2.44HH430 pKa = 6.38TMAVPTTTGHH440 pKa = 4.77IHH442 pKa = 6.76GGGGNDD448 pKa = 3.9GNHH451 pKa = 6.71GGHH454 pKa = 6.76GGMAMTFYY462 pKa = 10.77FGCNNVEE469 pKa = 4.16LLFSGLLINSPGEE482 pKa = 3.92MVGACIGVFLLAVLYY497 pKa = 10.14EE498 pKa = 4.05GLKK501 pKa = 9.13MGRR504 pKa = 11.84EE505 pKa = 3.9ALLRR509 pKa = 11.84RR510 pKa = 11.84SQVNVRR516 pKa = 11.84YY517 pKa = 10.52NSMPLPGADD526 pKa = 2.86GTVLMEE532 pKa = 3.68THH534 pKa = 5.73KK535 pKa = 9.83TVGQRR540 pKa = 11.84MLSPAHH546 pKa = 6.28FLQTLLHH553 pKa = 6.87IIQVVVSYY561 pKa = 10.47FLMLIFMTYY570 pKa = 9.8NAYY573 pKa = 10.68LCIAVAAGAGMGYY586 pKa = 10.54FLFSWRR592 pKa = 11.84KK593 pKa = 8.91AVVVDD598 pKa = 3.55ITEE601 pKa = 4.18HH602 pKa = 5.39CHH604 pKa = 5.81
MM1 pKa = 7.96IDD3 pKa = 4.76RR4 pKa = 11.84DD5 pKa = 4.14MIDD8 pKa = 4.84CDD10 pKa = 5.55VIDD13 pKa = 5.85CDD15 pKa = 5.73MIDD18 pKa = 4.12CDD20 pKa = 4.44VFDD23 pKa = 5.56RR24 pKa = 11.84DD25 pKa = 3.67VFDD28 pKa = 5.68RR29 pKa = 11.84DD30 pKa = 3.45VFDD33 pKa = 5.68RR34 pKa = 11.84DD35 pKa = 3.45VFDD38 pKa = 6.19RR39 pKa = 11.84DD40 pKa = 4.02VFDD43 pKa = 6.24CDD45 pKa = 5.14VFDD48 pKa = 6.49CDD50 pKa = 5.84VFDD53 pKa = 6.49CDD55 pKa = 5.84VFDD58 pKa = 5.74CDD60 pKa = 4.46VFDD63 pKa = 6.55RR64 pKa = 11.84DD65 pKa = 4.94VFDD68 pKa = 5.6CDD70 pKa = 3.53VFDD73 pKa = 6.55RR74 pKa = 11.84DD75 pKa = 4.94VFDD78 pKa = 5.6CDD80 pKa = 3.53VFDD83 pKa = 6.55RR84 pKa = 11.84DD85 pKa = 4.94VFDD88 pKa = 6.24CDD90 pKa = 5.14VFDD93 pKa = 5.74CDD95 pKa = 4.46VFDD98 pKa = 5.74RR99 pKa = 11.84DD100 pKa = 3.67VFDD103 pKa = 6.19RR104 pKa = 11.84DD105 pKa = 4.02VFDD108 pKa = 5.6CDD110 pKa = 3.53VFDD113 pKa = 6.55RR114 pKa = 11.84DD115 pKa = 4.94VFDD118 pKa = 6.24CDD120 pKa = 5.14VFDD123 pKa = 5.74CDD125 pKa = 4.46VFDD128 pKa = 6.55RR129 pKa = 11.84DD130 pKa = 4.94VFDD133 pKa = 5.6CDD135 pKa = 3.53VFDD138 pKa = 6.55RR139 pKa = 11.84DD140 pKa = 4.94VFDD143 pKa = 5.6CDD145 pKa = 3.53VFDD148 pKa = 6.55RR149 pKa = 11.84DD150 pKa = 4.94VFDD153 pKa = 5.6CDD155 pKa = 3.53VFDD158 pKa = 6.55RR159 pKa = 11.84DD160 pKa = 4.94VFDD163 pKa = 5.6CDD165 pKa = 3.53VFDD168 pKa = 5.74RR169 pKa = 11.84DD170 pKa = 3.67VFDD173 pKa = 5.68RR174 pKa = 11.84DD175 pKa = 3.45VFDD178 pKa = 5.68RR179 pKa = 11.84DD180 pKa = 3.45VFDD183 pKa = 6.19RR184 pKa = 11.84DD185 pKa = 4.02VFDD188 pKa = 6.24CDD190 pKa = 5.14VFDD193 pKa = 5.74CDD195 pKa = 4.46VFDD198 pKa = 6.55RR199 pKa = 11.84DD200 pKa = 4.94VFDD203 pKa = 6.24CDD205 pKa = 5.14VFDD208 pKa = 6.49CDD210 pKa = 5.84VFDD213 pKa = 5.74CDD215 pKa = 4.46VFDD218 pKa = 6.55RR219 pKa = 11.84DD220 pKa = 4.94VFDD223 pKa = 5.6CDD225 pKa = 3.49VFDD228 pKa = 6.49RR229 pKa = 11.84DD230 pKa = 5.38VIDD233 pKa = 5.75CDD235 pKa = 4.82VFDD238 pKa = 5.81CDD240 pKa = 5.13VIDD243 pKa = 5.91RR244 pKa = 11.84DD245 pKa = 4.62VFDD248 pKa = 5.66CDD250 pKa = 4.13VIDD253 pKa = 4.92RR254 pKa = 11.84DD255 pKa = 4.15VIDD258 pKa = 5.57RR259 pKa = 11.84DD260 pKa = 4.0VFDD263 pKa = 6.13CDD265 pKa = 4.55VFDD268 pKa = 5.74CNVIDD273 pKa = 6.05RR274 pKa = 11.84DD275 pKa = 4.14VFDD278 pKa = 5.66CDD280 pKa = 4.27VIDD283 pKa = 4.72RR284 pKa = 11.84DD285 pKa = 3.86VFDD288 pKa = 6.07RR289 pKa = 11.84DD290 pKa = 3.94VFDD293 pKa = 5.55CDD295 pKa = 4.27VIDD298 pKa = 5.91RR299 pKa = 11.84DD300 pKa = 4.79VFDD303 pKa = 6.04CDD305 pKa = 5.14VFDD308 pKa = 6.49CDD310 pKa = 5.84VFDD313 pKa = 6.49CDD315 pKa = 5.84VFDD318 pKa = 6.49CDD320 pKa = 5.84VFDD323 pKa = 6.49CDD325 pKa = 5.84VFDD328 pKa = 5.74CDD330 pKa = 4.46VFDD333 pKa = 5.74RR334 pKa = 11.84DD335 pKa = 3.67VFDD338 pKa = 6.19RR339 pKa = 11.84DD340 pKa = 4.02VFDD343 pKa = 6.24CDD345 pKa = 5.14VFDD348 pKa = 5.74CDD350 pKa = 4.26VFDD353 pKa = 5.78RR354 pKa = 11.84DD355 pKa = 4.18VIDD358 pKa = 4.79RR359 pKa = 11.84DD360 pKa = 3.69VFDD363 pKa = 5.72RR364 pKa = 11.84DD365 pKa = 3.45VFDD368 pKa = 5.68RR369 pKa = 11.84DD370 pKa = 3.45VFDD373 pKa = 5.68RR374 pKa = 11.84DD375 pKa = 3.45VFDD378 pKa = 5.68RR379 pKa = 11.84DD380 pKa = 3.45VFDD383 pKa = 5.68RR384 pKa = 11.84DD385 pKa = 3.45VFDD388 pKa = 6.19RR389 pKa = 11.84DD390 pKa = 4.02VFDD393 pKa = 6.24CDD395 pKa = 5.14VFDD398 pKa = 6.49CDD400 pKa = 5.84VFDD403 pKa = 5.6CDD405 pKa = 5.14VFDD408 pKa = 6.11FMNHH412 pKa = 6.56DD413 pKa = 4.18YY414 pKa = 10.18MNHH417 pKa = 4.38STMPMKK423 pKa = 10.3HH424 pKa = 6.36DD425 pKa = 3.4HH426 pKa = 6.93HH427 pKa = 7.38NDD429 pKa = 2.44HH430 pKa = 6.38TMAVPTTTGHH440 pKa = 4.77IHH442 pKa = 6.76GGGGNDD448 pKa = 3.9GNHH451 pKa = 6.71GGHH454 pKa = 6.76GGMAMTFYY462 pKa = 10.77FGCNNVEE469 pKa = 4.16LLFSGLLINSPGEE482 pKa = 3.92MVGACIGVFLLAVLYY497 pKa = 10.14EE498 pKa = 4.05GLKK501 pKa = 9.13MGRR504 pKa = 11.84EE505 pKa = 3.9ALLRR509 pKa = 11.84RR510 pKa = 11.84SQVNVRR516 pKa = 11.84YY517 pKa = 10.52NSMPLPGADD526 pKa = 2.86GTVLMEE532 pKa = 3.68THH534 pKa = 5.73KK535 pKa = 9.83TVGQRR540 pKa = 11.84MLSPAHH546 pKa = 6.28FLQTLLHH553 pKa = 6.87IIQVVVSYY561 pKa = 10.47FLMLIFMTYY570 pKa = 9.8NAYY573 pKa = 10.68LCIAVAAGAGMGYY586 pKa = 10.54FLFSWRR592 pKa = 11.84KK593 pKa = 8.91AVVVDD598 pKa = 3.55ITEE601 pKa = 4.18HH602 pKa = 5.39CHH604 pKa = 5.81
Molecular weight: 70.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4U5V576|A0A4U5V576_COLLU Upstream stimulatory factor 2 OS=Collichthys lucidus OX=240159 GN=D9C73_016529 PE=4 SV=1
MM1 pKa = 7.52LNATRR6 pKa = 11.84MLNATRR12 pKa = 11.84MLNATRR18 pKa = 11.84MLNATRR24 pKa = 11.84MLNGTRR30 pKa = 11.84MLNAATRR37 pKa = 11.84MLNAATRR44 pKa = 11.84MLNAATRR51 pKa = 11.84MLNATRR57 pKa = 11.84MLNATRR63 pKa = 11.84MLNATRR69 pKa = 11.84MLNATRR75 pKa = 11.84MLNATRR81 pKa = 11.84MLNATRR87 pKa = 11.84MLNATRR93 pKa = 11.84MLNATHH99 pKa = 6.6MLNAATRR106 pKa = 11.84MLNAATRR113 pKa = 11.84MLNATRR119 pKa = 11.84MLNAATRR126 pKa = 11.84MLNATRR132 pKa = 11.84MLNAATRR139 pKa = 11.84MLNATRR145 pKa = 11.84MLNAATRR152 pKa = 11.84MLNATRR158 pKa = 11.84MLNAATRR165 pKa = 11.84MLNATRR171 pKa = 11.84MLNAATRR178 pKa = 11.84MLNATRR184 pKa = 11.84MLNARR189 pKa = 11.84VCC191 pKa = 3.72
MM1 pKa = 7.52LNATRR6 pKa = 11.84MLNATRR12 pKa = 11.84MLNATRR18 pKa = 11.84MLNATRR24 pKa = 11.84MLNGTRR30 pKa = 11.84MLNAATRR37 pKa = 11.84MLNAATRR44 pKa = 11.84MLNAATRR51 pKa = 11.84MLNATRR57 pKa = 11.84MLNATRR63 pKa = 11.84MLNATRR69 pKa = 11.84MLNATRR75 pKa = 11.84MLNATRR81 pKa = 11.84MLNATRR87 pKa = 11.84MLNATRR93 pKa = 11.84MLNATHH99 pKa = 6.6MLNAATRR106 pKa = 11.84MLNAATRR113 pKa = 11.84MLNATRR119 pKa = 11.84MLNAATRR126 pKa = 11.84MLNATRR132 pKa = 11.84MLNAATRR139 pKa = 11.84MLNATRR145 pKa = 11.84MLNAATRR152 pKa = 11.84MLNATRR158 pKa = 11.84MLNAATRR165 pKa = 11.84MLNATRR171 pKa = 11.84MLNAATRR178 pKa = 11.84MLNATRR184 pKa = 11.84MLNARR189 pKa = 11.84VCC191 pKa = 3.72
Molecular weight: 21.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
15347301 |
49 |
38162 |
564.1 |
62.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.629 ± 0.014 | 2.195 ± 0.011 |
5.257 ± 0.011 | 6.964 ± 0.024 |
3.439 ± 0.01 | 6.322 ± 0.021 |
2.679 ± 0.009 | 4.217 ± 0.013 |
5.553 ± 0.02 | 9.421 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.387 ± 0.008 | 3.771 ± 0.011 |
5.796 ± 0.021 | 4.813 ± 0.015 |
5.812 ± 0.015 | 8.786 ± 0.021 |
5.81 ± 0.019 | 6.354 ± 0.014 |
1.127 ± 0.006 | 2.667 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
