
Ceratocystis fimbriata CBS 114723
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Microascales; Ceratocystidaceae; Ceratocystis; Ceratocystis fimbriata
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
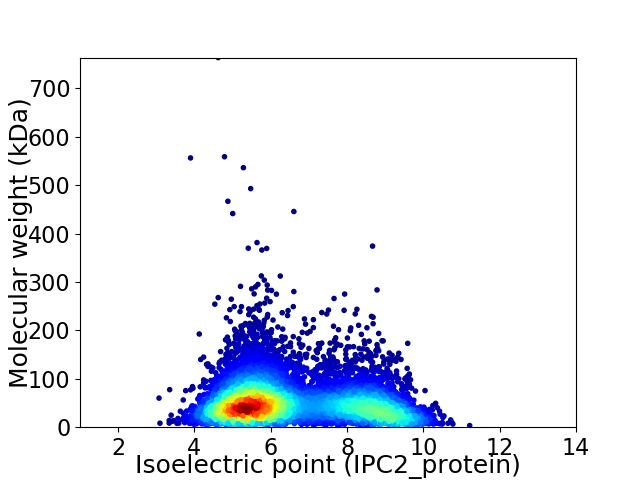
Virtual 2D-PAGE plot for 7264 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
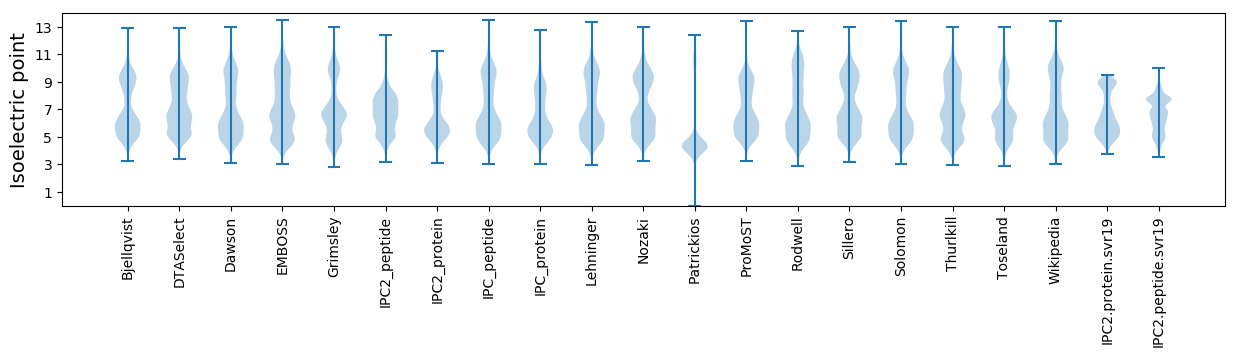
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2C5XF84|A0A2C5XF84_9PEZI Sulfate permease 2 OS=Ceratocystis fimbriata CBS 114723 OX=1035309 GN=cys-14_1 PE=3 SV=1
MM1 pKa = 7.17KK2 pKa = 9.95TFATIVASASLVAAQGYY19 pKa = 6.88MTGCSIGNQNFKK31 pKa = 10.62FSNVSSTDD39 pKa = 2.82SYY41 pKa = 11.95SNVNADD47 pKa = 4.1RR48 pKa = 11.84ISRR51 pKa = 11.84YY52 pKa = 5.16TTGSTGVKK60 pKa = 10.14DD61 pKa = 3.79VTSLDD66 pKa = 3.9MQCGATGGKK75 pKa = 7.88GTQGAPEE82 pKa = 4.18SAKK85 pKa = 10.08VAAGEE90 pKa = 4.22SVQMHH95 pKa = 5.71WSKK98 pKa = 8.04WTKK101 pKa = 9.14VNKK104 pKa = 10.2GPTVTYY110 pKa = 9.02MAKK113 pKa = 10.44CPDD116 pKa = 4.68AGCQDD121 pKa = 4.33YY122 pKa = 11.33VPGTDD127 pKa = 3.57AVWFKK132 pKa = 10.68IQEE135 pKa = 4.41AGPTASGQWPSTSLGTSGSQDD156 pKa = 2.97TYY158 pKa = 11.03TIPEE162 pKa = 4.24CLASGNYY169 pKa = 9.23LVRR172 pKa = 11.84HH173 pKa = 5.54EE174 pKa = 5.17AISLGGSAQFYY185 pKa = 10.03IGCHH189 pKa = 4.21QLEE192 pKa = 4.51VSGSGTTNPTDD203 pKa = 3.28LVSFPGAYY211 pKa = 9.35KK212 pKa = 10.74SSDD215 pKa = 3.18SGIKK219 pKa = 9.82FDD221 pKa = 5.33PNTMPPSSYY230 pKa = 9.66TVPGPSVFTCGGSSGSTPSAAGSSSGPKK258 pKa = 9.68PSTGPSSAVSSNSPVGPSNVPMGTGNTPKK287 pKa = 9.52VTGNLPKK294 pKa = 10.37GTGNVPLGTGSVGSNGGSATTPASGYY320 pKa = 7.07QTIASVSGSASAAATTPAPGGEE342 pKa = 4.27YY343 pKa = 10.32GAEE346 pKa = 4.15DD347 pKa = 3.93VSSVPVRR354 pKa = 11.84GTGSATSASSPVTTSGPEE372 pKa = 3.84DD373 pKa = 3.83TEE375 pKa = 4.36DD376 pKa = 4.31CPVEE380 pKa = 4.39NDD382 pKa = 3.96TVPASPTSSRR392 pKa = 11.84VPLSGTNSTIPDD404 pKa = 3.91GGSTTVGSSPLTTGSTGSGSGSDD427 pKa = 3.89YY428 pKa = 11.25EE429 pKa = 5.05SGDD432 pKa = 3.73DD433 pKa = 3.67EE434 pKa = 5.88CPVEE438 pKa = 5.66DD439 pKa = 5.67GEE441 pKa = 4.57TTPAGSATTASSPASTGAGSTTPVEE466 pKa = 4.71SDD468 pKa = 4.11DD469 pKa = 4.13EE470 pKa = 4.62CPADD474 pKa = 4.43DD475 pKa = 5.41SEE477 pKa = 4.59EE478 pKa = 4.3TTSNSSPTTPEE489 pKa = 3.67TKK491 pKa = 9.99TPTGSSPSSSDD502 pKa = 2.76SKK504 pKa = 10.09TPSDD508 pKa = 4.09SSPSTSDD515 pKa = 2.71SKK517 pKa = 10.24TPTSSSLSSSGSNTPSGSSPSSYY540 pKa = 10.81GSKK543 pKa = 9.06TPSGSSPSTSDD554 pKa = 2.74SKK556 pKa = 10.21TPTSSSPSSSSSSSSTGSDD575 pKa = 3.12SYY577 pKa = 10.88STSNTEE583 pKa = 3.95STSTNTGKK591 pKa = 10.21NSYY594 pKa = 8.17STEE597 pKa = 3.85SGYY600 pKa = 11.56GSGTGSGTGTGSGSTTNPSTPKK622 pKa = 10.6SSTTSAASPQSPQTSSSASPEE643 pKa = 3.98NDD645 pKa = 3.5DD646 pKa = 3.64EE647 pKa = 5.44CPAEE651 pKa = 4.43DD652 pKa = 4.07EE653 pKa = 4.53EE654 pKa = 5.46SVEE657 pKa = 5.11EE658 pKa = 4.94DD659 pKa = 3.9DD660 pKa = 4.32EE661 pKa = 5.06CPAEE665 pKa = 4.17EE666 pKa = 5.25DD667 pKa = 3.95EE668 pKa = 4.92PATTSPVEE676 pKa = 4.15TQNDD680 pKa = 3.99DD681 pKa = 3.02QCAADD686 pKa = 5.06DD687 pKa = 4.92DD688 pKa = 4.81EE689 pKa = 4.8EE690 pKa = 4.63TTSNSSPTTPTTTEE704 pKa = 4.29EE705 pKa = 5.25DD706 pKa = 4.07DD707 pKa = 4.28DD708 pKa = 4.59ACPADD713 pKa = 4.07EE714 pKa = 5.8DD715 pKa = 4.61EE716 pKa = 4.97PTTTTPTSPSSTSGSASTSGTSTSSASPEE745 pKa = 4.13TEE747 pKa = 3.72EE748 pKa = 4.15EE749 pKa = 4.53CPVEE753 pKa = 4.03YY754 pKa = 10.49EE755 pKa = 3.94RR756 pKa = 11.84DD757 pKa = 3.33LSYY760 pKa = 11.46VEE762 pKa = 4.94EE763 pKa = 4.22NLPRR767 pKa = 11.84RR768 pKa = 11.84HH769 pKa = 5.86WDD771 pKa = 3.11RR772 pKa = 11.84RR773 pKa = 11.84SWW775 pKa = 3.2
MM1 pKa = 7.17KK2 pKa = 9.95TFATIVASASLVAAQGYY19 pKa = 6.88MTGCSIGNQNFKK31 pKa = 10.62FSNVSSTDD39 pKa = 2.82SYY41 pKa = 11.95SNVNADD47 pKa = 4.1RR48 pKa = 11.84ISRR51 pKa = 11.84YY52 pKa = 5.16TTGSTGVKK60 pKa = 10.14DD61 pKa = 3.79VTSLDD66 pKa = 3.9MQCGATGGKK75 pKa = 7.88GTQGAPEE82 pKa = 4.18SAKK85 pKa = 10.08VAAGEE90 pKa = 4.22SVQMHH95 pKa = 5.71WSKK98 pKa = 8.04WTKK101 pKa = 9.14VNKK104 pKa = 10.2GPTVTYY110 pKa = 9.02MAKK113 pKa = 10.44CPDD116 pKa = 4.68AGCQDD121 pKa = 4.33YY122 pKa = 11.33VPGTDD127 pKa = 3.57AVWFKK132 pKa = 10.68IQEE135 pKa = 4.41AGPTASGQWPSTSLGTSGSQDD156 pKa = 2.97TYY158 pKa = 11.03TIPEE162 pKa = 4.24CLASGNYY169 pKa = 9.23LVRR172 pKa = 11.84HH173 pKa = 5.54EE174 pKa = 5.17AISLGGSAQFYY185 pKa = 10.03IGCHH189 pKa = 4.21QLEE192 pKa = 4.51VSGSGTTNPTDD203 pKa = 3.28LVSFPGAYY211 pKa = 9.35KK212 pKa = 10.74SSDD215 pKa = 3.18SGIKK219 pKa = 9.82FDD221 pKa = 5.33PNTMPPSSYY230 pKa = 9.66TVPGPSVFTCGGSSGSTPSAAGSSSGPKK258 pKa = 9.68PSTGPSSAVSSNSPVGPSNVPMGTGNTPKK287 pKa = 9.52VTGNLPKK294 pKa = 10.37GTGNVPLGTGSVGSNGGSATTPASGYY320 pKa = 7.07QTIASVSGSASAAATTPAPGGEE342 pKa = 4.27YY343 pKa = 10.32GAEE346 pKa = 4.15DD347 pKa = 3.93VSSVPVRR354 pKa = 11.84GTGSATSASSPVTTSGPEE372 pKa = 3.84DD373 pKa = 3.83TEE375 pKa = 4.36DD376 pKa = 4.31CPVEE380 pKa = 4.39NDD382 pKa = 3.96TVPASPTSSRR392 pKa = 11.84VPLSGTNSTIPDD404 pKa = 3.91GGSTTVGSSPLTTGSTGSGSGSDD427 pKa = 3.89YY428 pKa = 11.25EE429 pKa = 5.05SGDD432 pKa = 3.73DD433 pKa = 3.67EE434 pKa = 5.88CPVEE438 pKa = 5.66DD439 pKa = 5.67GEE441 pKa = 4.57TTPAGSATTASSPASTGAGSTTPVEE466 pKa = 4.71SDD468 pKa = 4.11DD469 pKa = 4.13EE470 pKa = 4.62CPADD474 pKa = 4.43DD475 pKa = 5.41SEE477 pKa = 4.59EE478 pKa = 4.3TTSNSSPTTPEE489 pKa = 3.67TKK491 pKa = 9.99TPTGSSPSSSDD502 pKa = 2.76SKK504 pKa = 10.09TPSDD508 pKa = 4.09SSPSTSDD515 pKa = 2.71SKK517 pKa = 10.24TPTSSSLSSSGSNTPSGSSPSSYY540 pKa = 10.81GSKK543 pKa = 9.06TPSGSSPSTSDD554 pKa = 2.74SKK556 pKa = 10.21TPTSSSPSSSSSSSSTGSDD575 pKa = 3.12SYY577 pKa = 10.88STSNTEE583 pKa = 3.95STSTNTGKK591 pKa = 10.21NSYY594 pKa = 8.17STEE597 pKa = 3.85SGYY600 pKa = 11.56GSGTGSGTGTGSGSTTNPSTPKK622 pKa = 10.6SSTTSAASPQSPQTSSSASPEE643 pKa = 3.98NDD645 pKa = 3.5DD646 pKa = 3.64EE647 pKa = 5.44CPAEE651 pKa = 4.43DD652 pKa = 4.07EE653 pKa = 4.53EE654 pKa = 5.46SVEE657 pKa = 5.11EE658 pKa = 4.94DD659 pKa = 3.9DD660 pKa = 4.32EE661 pKa = 5.06CPAEE665 pKa = 4.17EE666 pKa = 5.25DD667 pKa = 3.95EE668 pKa = 4.92PATTSPVEE676 pKa = 4.15TQNDD680 pKa = 3.99DD681 pKa = 3.02QCAADD686 pKa = 5.06DD687 pKa = 4.92DD688 pKa = 4.81EE689 pKa = 4.8EE690 pKa = 4.63TTSNSSPTTPTTTEE704 pKa = 4.29EE705 pKa = 5.25DD706 pKa = 4.07DD707 pKa = 4.28DD708 pKa = 4.59ACPADD713 pKa = 4.07EE714 pKa = 5.8DD715 pKa = 4.61EE716 pKa = 4.97PTTTTPTSPSSTSGSASTSGTSTSSASPEE745 pKa = 4.13TEE747 pKa = 3.72EE748 pKa = 4.15EE749 pKa = 4.53CPVEE753 pKa = 4.03YY754 pKa = 10.49EE755 pKa = 3.94RR756 pKa = 11.84DD757 pKa = 3.33LSYY760 pKa = 11.46VEE762 pKa = 4.94EE763 pKa = 4.22NLPRR767 pKa = 11.84RR768 pKa = 11.84HH769 pKa = 5.86WDD771 pKa = 3.11RR772 pKa = 11.84RR773 pKa = 11.84SWW775 pKa = 3.2
Molecular weight: 77.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2C5WXN3|A0A2C5WXN3_9PEZI Uncharacterized protein OS=Ceratocystis fimbriata CBS 114723 OX=1035309 GN=CFIMG_007429RA00001 PE=4 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.09WRR6 pKa = 11.84KK7 pKa = 8.87KK8 pKa = 8.73RR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.94RR15 pKa = 11.84KK16 pKa = 7.8RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.6MRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 11.11
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.09WRR6 pKa = 11.84KK7 pKa = 8.87KK8 pKa = 8.73RR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.94RR15 pKa = 11.84KK16 pKa = 7.8RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.6MRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 11.11
Molecular weight: 3.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3783215 |
17 |
7066 |
520.8 |
57.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.052 ± 0.035 | 1.151 ± 0.009 |
5.711 ± 0.022 | 5.971 ± 0.033 |
3.431 ± 0.016 | 6.37 ± 0.025 |
2.436 ± 0.014 | 4.822 ± 0.02 |
4.871 ± 0.024 | 8.468 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.394 ± 0.012 | 3.848 ± 0.015 |
6.269 ± 0.033 | 4.09 ± 0.023 |
5.969 ± 0.028 | 9.263 ± 0.038 |
6.174 ± 0.025 | 5.943 ± 0.022 |
1.228 ± 0.01 | 2.53 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
