
Trematomus bernacchii polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
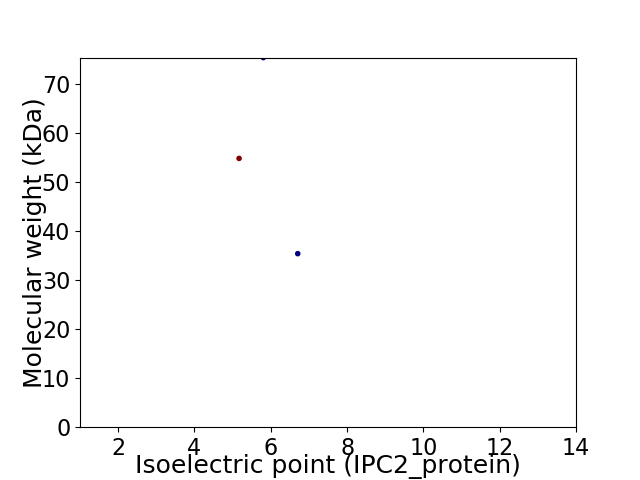
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U9K629|A0A2U9K629_9POLY VP2 OS=Trematomus bernacchii polyomavirus 1 OX=2218588 GN=VP2 PE=4 SV=1
MM1 pKa = 7.38GLVISVVAGISAFFAEE17 pKa = 4.74LTVAAAMAGTSAEE30 pKa = 3.93AVTASFEE37 pKa = 4.22LSVVLGEE44 pKa = 4.26SAEE47 pKa = 3.99DD48 pKa = 3.49VMAGEE53 pKa = 4.72EE54 pKa = 4.06FTLFSGEE61 pKa = 3.9TAEE64 pKa = 4.37AEE66 pKa = 4.67GYY68 pKa = 10.1VDD70 pKa = 4.42EE71 pKa = 5.64GAINDD76 pKa = 3.42AWNWSPIEE84 pKa = 3.97GTTRR88 pKa = 11.84STFSGVARR96 pKa = 11.84SRR98 pKa = 11.84LAQTVGSGIVAASVIGAVAGTQLEE122 pKa = 4.38GDD124 pKa = 3.43QGLDD128 pKa = 2.94MVRR131 pKa = 11.84AVFNPEE137 pKa = 3.34SSGDD141 pKa = 3.67HH142 pKa = 5.31TVSDD146 pKa = 3.48TMEE149 pKa = 5.78RR150 pKa = 11.84IMTPEE155 pKa = 3.84EE156 pKa = 4.61YY157 pKa = 9.61IDD159 pKa = 3.55NMQILSEE166 pKa = 4.29EE167 pKa = 4.45YY168 pKa = 10.6SNGMLSRR175 pKa = 11.84DD176 pKa = 3.71LPLSVAVADD185 pKa = 3.72YY186 pKa = 10.56SAAIGRR192 pKa = 11.84SGRR195 pKa = 11.84LAHH198 pKa = 6.55RR199 pKa = 11.84TRR201 pKa = 11.84EE202 pKa = 3.95TMGADD207 pKa = 3.59KK208 pKa = 11.02SDD210 pKa = 3.91LLIFSAMKK218 pKa = 10.4SEE220 pKa = 4.22PKK222 pKa = 10.17VGSWTLPGADD232 pKa = 4.91GIPITRR238 pKa = 11.84AAWNRR243 pKa = 11.84YY244 pKa = 5.2TMGYY248 pKa = 8.53YY249 pKa = 10.2HH250 pKa = 6.81GTAAQMRR257 pKa = 11.84EE258 pKa = 3.98MLAHH262 pKa = 6.66GEE264 pKa = 4.25VLRR267 pKa = 11.84QAGQNIWPLFQRR279 pKa = 11.84AAHH282 pKa = 6.64LAAAPVAVALDD293 pKa = 3.67AALDD297 pKa = 3.92VPQDD301 pKa = 3.44GVRR304 pKa = 11.84QQLRR308 pKa = 11.84GIGRR312 pKa = 11.84RR313 pKa = 11.84AAHH316 pKa = 6.85RR317 pKa = 11.84LAGDD321 pKa = 3.41AGVAVAQLGDD331 pKa = 3.51RR332 pKa = 11.84VANALVAGGNRR343 pKa = 11.84AGHH346 pKa = 5.24QAVEE350 pKa = 4.25TVIAALTYY358 pKa = 10.58GVTEE362 pKa = 4.34YY363 pKa = 10.77FSSTSSPKK371 pKa = 9.92PGPDD375 pKa = 3.4AIEE378 pKa = 5.23IEE380 pKa = 4.95DD381 pKa = 5.12DD382 pKa = 3.63PDD384 pKa = 3.57LPKK387 pKa = 10.59EE388 pKa = 3.98IEE390 pKa = 4.09VDD392 pKa = 2.65GRR394 pKa = 11.84TYY396 pKa = 11.08VKK398 pKa = 10.54EE399 pKa = 3.87KK400 pKa = 11.0ADD402 pKa = 3.78LHH404 pKa = 5.94TVTHH408 pKa = 6.82PEE410 pKa = 3.49GTYY413 pKa = 8.94THH415 pKa = 7.07TEE417 pKa = 4.01VHH419 pKa = 5.99WYY421 pKa = 9.69EE422 pKa = 3.92PTEE425 pKa = 4.27RR426 pKa = 11.84PQIAPGYY433 pKa = 7.93DD434 pKa = 2.93TAFKK438 pKa = 10.12YY439 pKa = 10.74AEE441 pKa = 4.43PRR443 pKa = 11.84SPDD446 pKa = 3.06QMLALTEE453 pKa = 3.89VLGYY457 pKa = 9.59FVHH460 pKa = 7.21GYY462 pKa = 10.64DD463 pKa = 3.19ITPPLTTSQRR473 pKa = 11.84LILSALTLQVARR485 pKa = 11.84RR486 pKa = 11.84HH487 pKa = 5.42ASRR490 pKa = 11.84KK491 pKa = 8.13RR492 pKa = 11.84RR493 pKa = 11.84IRR495 pKa = 11.84GTEE498 pKa = 3.3HH499 pKa = 7.24RR500 pKa = 11.84SRR502 pKa = 11.84SKK504 pKa = 10.67RR505 pKa = 11.84KK506 pKa = 9.72RR507 pKa = 11.84EE508 pKa = 3.82AA509 pKa = 3.11
MM1 pKa = 7.38GLVISVVAGISAFFAEE17 pKa = 4.74LTVAAAMAGTSAEE30 pKa = 3.93AVTASFEE37 pKa = 4.22LSVVLGEE44 pKa = 4.26SAEE47 pKa = 3.99DD48 pKa = 3.49VMAGEE53 pKa = 4.72EE54 pKa = 4.06FTLFSGEE61 pKa = 3.9TAEE64 pKa = 4.37AEE66 pKa = 4.67GYY68 pKa = 10.1VDD70 pKa = 4.42EE71 pKa = 5.64GAINDD76 pKa = 3.42AWNWSPIEE84 pKa = 3.97GTTRR88 pKa = 11.84STFSGVARR96 pKa = 11.84SRR98 pKa = 11.84LAQTVGSGIVAASVIGAVAGTQLEE122 pKa = 4.38GDD124 pKa = 3.43QGLDD128 pKa = 2.94MVRR131 pKa = 11.84AVFNPEE137 pKa = 3.34SSGDD141 pKa = 3.67HH142 pKa = 5.31TVSDD146 pKa = 3.48TMEE149 pKa = 5.78RR150 pKa = 11.84IMTPEE155 pKa = 3.84EE156 pKa = 4.61YY157 pKa = 9.61IDD159 pKa = 3.55NMQILSEE166 pKa = 4.29EE167 pKa = 4.45YY168 pKa = 10.6SNGMLSRR175 pKa = 11.84DD176 pKa = 3.71LPLSVAVADD185 pKa = 3.72YY186 pKa = 10.56SAAIGRR192 pKa = 11.84SGRR195 pKa = 11.84LAHH198 pKa = 6.55RR199 pKa = 11.84TRR201 pKa = 11.84EE202 pKa = 3.95TMGADD207 pKa = 3.59KK208 pKa = 11.02SDD210 pKa = 3.91LLIFSAMKK218 pKa = 10.4SEE220 pKa = 4.22PKK222 pKa = 10.17VGSWTLPGADD232 pKa = 4.91GIPITRR238 pKa = 11.84AAWNRR243 pKa = 11.84YY244 pKa = 5.2TMGYY248 pKa = 8.53YY249 pKa = 10.2HH250 pKa = 6.81GTAAQMRR257 pKa = 11.84EE258 pKa = 3.98MLAHH262 pKa = 6.66GEE264 pKa = 4.25VLRR267 pKa = 11.84QAGQNIWPLFQRR279 pKa = 11.84AAHH282 pKa = 6.64LAAAPVAVALDD293 pKa = 3.67AALDD297 pKa = 3.92VPQDD301 pKa = 3.44GVRR304 pKa = 11.84QQLRR308 pKa = 11.84GIGRR312 pKa = 11.84RR313 pKa = 11.84AAHH316 pKa = 6.85RR317 pKa = 11.84LAGDD321 pKa = 3.41AGVAVAQLGDD331 pKa = 3.51RR332 pKa = 11.84VANALVAGGNRR343 pKa = 11.84AGHH346 pKa = 5.24QAVEE350 pKa = 4.25TVIAALTYY358 pKa = 10.58GVTEE362 pKa = 4.34YY363 pKa = 10.77FSSTSSPKK371 pKa = 9.92PGPDD375 pKa = 3.4AIEE378 pKa = 5.23IEE380 pKa = 4.95DD381 pKa = 5.12DD382 pKa = 3.63PDD384 pKa = 3.57LPKK387 pKa = 10.59EE388 pKa = 3.98IEE390 pKa = 4.09VDD392 pKa = 2.65GRR394 pKa = 11.84TYY396 pKa = 11.08VKK398 pKa = 10.54EE399 pKa = 3.87KK400 pKa = 11.0ADD402 pKa = 3.78LHH404 pKa = 5.94TVTHH408 pKa = 6.82PEE410 pKa = 3.49GTYY413 pKa = 8.94THH415 pKa = 7.07TEE417 pKa = 4.01VHH419 pKa = 5.99WYY421 pKa = 9.69EE422 pKa = 3.92PTEE425 pKa = 4.27RR426 pKa = 11.84PQIAPGYY433 pKa = 7.93DD434 pKa = 2.93TAFKK438 pKa = 10.12YY439 pKa = 10.74AEE441 pKa = 4.43PRR443 pKa = 11.84SPDD446 pKa = 3.06QMLALTEE453 pKa = 3.89VLGYY457 pKa = 9.59FVHH460 pKa = 7.21GYY462 pKa = 10.64DD463 pKa = 3.19ITPPLTTSQRR473 pKa = 11.84LILSALTLQVARR485 pKa = 11.84RR486 pKa = 11.84HH487 pKa = 5.42ASRR490 pKa = 11.84KK491 pKa = 8.13RR492 pKa = 11.84RR493 pKa = 11.84IRR495 pKa = 11.84GTEE498 pKa = 3.3HH499 pKa = 7.24RR500 pKa = 11.84SRR502 pKa = 11.84SKK504 pKa = 10.67RR505 pKa = 11.84KK506 pKa = 9.72RR507 pKa = 11.84EE508 pKa = 3.82AA509 pKa = 3.11
Molecular weight: 54.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U9K628|A0A2U9K628_9POLY Large T antigen OS=Trematomus bernacchii polyomavirus 1 OX=2218588 PE=4 SV=1
MM1 pKa = 7.43PVVKK5 pKa = 10.34GGYY8 pKa = 8.35EE9 pKa = 3.87VLNIVADD16 pKa = 4.19LNEE19 pKa = 4.23SEE21 pKa = 5.34RR22 pKa = 11.84LDD24 pKa = 3.62INCYY28 pKa = 9.42LAPEE32 pKa = 5.46AYY34 pKa = 7.92TQAVGSNKK42 pKa = 9.61SPKK45 pKa = 8.99WLTSAIDD52 pKa = 3.69LPKK55 pKa = 10.38IPVAAKK61 pKa = 10.09GNKK64 pKa = 8.29IKK66 pKa = 10.1IWEE69 pKa = 4.48CYY71 pKa = 9.04KK72 pKa = 10.85CSTGVLTGDD81 pKa = 3.61PNHH84 pKa = 6.14VNEE87 pKa = 5.18IPSQSPYY94 pKa = 10.43NGPSRR99 pKa = 11.84SSWCVSGLPMEE110 pKa = 5.83GFGKK114 pKa = 9.73WPFPEE119 pKa = 5.62APPQPSILMKK129 pKa = 10.59NDD131 pKa = 4.16LYY133 pKa = 11.33DD134 pKa = 3.81STDD137 pKa = 3.78FTPNPWMEE145 pKa = 3.66NAKK148 pKa = 10.64YY149 pKa = 10.14YY150 pKa = 10.47VSKK153 pKa = 11.1QMGTQSYY160 pKa = 10.32AAGDD164 pKa = 3.89NNSSVVLLADD174 pKa = 3.77ADD176 pKa = 4.3GFGIMCPQGSCFISALDD193 pKa = 4.22FFTQYY198 pKa = 10.9ISGKK202 pKa = 7.47TVFQCYY208 pKa = 9.53QIPRR212 pKa = 11.84YY213 pKa = 8.96FKK215 pKa = 10.55LYY217 pKa = 10.09FRR219 pKa = 11.84QRR221 pKa = 11.84WVKK224 pKa = 10.61SPVVLMDD231 pKa = 5.24LFTQMVNQQVQGYY244 pKa = 9.54AGEE247 pKa = 4.27TGNIEE252 pKa = 4.3HH253 pKa = 7.11VSLAWGKK260 pKa = 10.5RR261 pKa = 11.84DD262 pKa = 3.72LVPGGLLGNTLNKK275 pKa = 10.3LGAGTSTSTTMDD287 pKa = 3.33TEE289 pKa = 4.39EE290 pKa = 4.62PPALPVKK297 pKa = 10.03LAKK300 pKa = 10.49SQVGSYY306 pKa = 9.11KK307 pKa = 10.35QPSASKK313 pKa = 9.69IPPGVTITNKK323 pKa = 10.31
MM1 pKa = 7.43PVVKK5 pKa = 10.34GGYY8 pKa = 8.35EE9 pKa = 3.87VLNIVADD16 pKa = 4.19LNEE19 pKa = 4.23SEE21 pKa = 5.34RR22 pKa = 11.84LDD24 pKa = 3.62INCYY28 pKa = 9.42LAPEE32 pKa = 5.46AYY34 pKa = 7.92TQAVGSNKK42 pKa = 9.61SPKK45 pKa = 8.99WLTSAIDD52 pKa = 3.69LPKK55 pKa = 10.38IPVAAKK61 pKa = 10.09GNKK64 pKa = 8.29IKK66 pKa = 10.1IWEE69 pKa = 4.48CYY71 pKa = 9.04KK72 pKa = 10.85CSTGVLTGDD81 pKa = 3.61PNHH84 pKa = 6.14VNEE87 pKa = 5.18IPSQSPYY94 pKa = 10.43NGPSRR99 pKa = 11.84SSWCVSGLPMEE110 pKa = 5.83GFGKK114 pKa = 9.73WPFPEE119 pKa = 5.62APPQPSILMKK129 pKa = 10.59NDD131 pKa = 4.16LYY133 pKa = 11.33DD134 pKa = 3.81STDD137 pKa = 3.78FTPNPWMEE145 pKa = 3.66NAKK148 pKa = 10.64YY149 pKa = 10.14YY150 pKa = 10.47VSKK153 pKa = 11.1QMGTQSYY160 pKa = 10.32AAGDD164 pKa = 3.89NNSSVVLLADD174 pKa = 3.77ADD176 pKa = 4.3GFGIMCPQGSCFISALDD193 pKa = 4.22FFTQYY198 pKa = 10.9ISGKK202 pKa = 7.47TVFQCYY208 pKa = 9.53QIPRR212 pKa = 11.84YY213 pKa = 8.96FKK215 pKa = 10.55LYY217 pKa = 10.09FRR219 pKa = 11.84QRR221 pKa = 11.84WVKK224 pKa = 10.61SPVVLMDD231 pKa = 5.24LFTQMVNQQVQGYY244 pKa = 9.54AGEE247 pKa = 4.27TGNIEE252 pKa = 4.3HH253 pKa = 7.11VSLAWGKK260 pKa = 10.5RR261 pKa = 11.84DD262 pKa = 3.72LVPGGLLGNTLNKK275 pKa = 10.3LGAGTSTSTTMDD287 pKa = 3.33TEE289 pKa = 4.39EE290 pKa = 4.62PPALPVKK297 pKa = 10.03LAKK300 pKa = 10.49SQVGSYY306 pKa = 9.11KK307 pKa = 10.35QPSASKK313 pKa = 9.69IPPGVTITNKK323 pKa = 10.31
Molecular weight: 35.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1504 |
323 |
672 |
501.3 |
55.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.71 ± 1.925 | 2.128 ± 0.932 |
5.718 ± 0.556 | 6.316 ± 0.696 |
3.856 ± 0.737 | 7.779 ± 0.576 |
2.194 ± 0.462 | 4.322 ± 0.163 |
5.319 ± 1.258 | 7.713 ± 0.25 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.593 ± 0.112 | 3.457 ± 0.763 |
5.718 ± 0.79 | 3.989 ± 0.32 |
5.053 ± 1.034 | 7.513 ± 0.34 |
5.851 ± 0.63 | 7.181 ± 0.204 |
1.263 ± 0.28 | 3.324 ± 0.402 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
