
Brazilian marseillevirus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Pimascovirales; Marseilleviridae; Marseillevirus; unclassified Marseillevirus
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
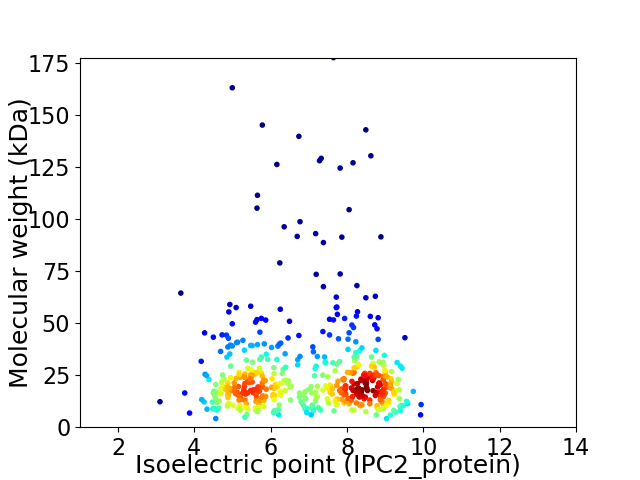
Virtual 2D-PAGE plot for 491 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A142CKN4|A0A142CKN4_9VIRU DUF5858 domain-containing protein OS=Brazilian marseillevirus OX=1813599 PE=4 SV=1
MM1 pKa = 7.38QNTTNFFRR9 pKa = 11.84DD10 pKa = 3.14ISLVGSVTSLPPVLSRR26 pKa = 11.84GTLVYY31 pKa = 10.64PEE33 pKa = 5.22DD34 pKa = 3.8SSNLYY39 pKa = 9.91ISNGSQWVIAGGDD52 pKa = 3.71VGPLAAQVAQNTADD66 pKa = 3.3IGVLQGDD73 pKa = 3.92VSTLQGEE80 pKa = 4.8VATNTTDD87 pKa = 3.38IAGLQTDD94 pKa = 3.94VSTLQGQVSSLSTDD108 pKa = 3.05VSALQTQVATNTTDD122 pKa = 2.61IGTLQGDD129 pKa = 3.76VSTLQGEE136 pKa = 4.85VATNTADD143 pKa = 3.34ISTLQGDD150 pKa = 4.07VSTLQTQVNANTLDD164 pKa = 3.27ISALQTQVATNTTDD178 pKa = 2.61IGTLQGDD185 pKa = 3.76VSTLQGEE192 pKa = 4.8VATNTSNITTLQGQVATNTLDD213 pKa = 3.05IGTLQTDD220 pKa = 3.6VTTLQGDD227 pKa = 3.79VSTLQTQVGTNTSNITTLQGQVATNTTDD255 pKa = 2.86ISALQTQVSTNTTNIGTLQGDD276 pKa = 3.84VSTLQTQVTTNTLDD290 pKa = 2.96IGNLQTDD297 pKa = 4.08VSTLQTDD304 pKa = 4.24VAQNTLDD311 pKa = 3.23ISGLQGQVATNTTNIATNTSNISTLQGQTATNTTNITTLQGQVSTNTSDD360 pKa = 3.16ISTLQTQVSTNTADD374 pKa = 2.92IGTLQGQVATNTTNIATNTSNISALQGQVSTNTSNISTLQTTKK417 pKa = 10.58EE418 pKa = 4.04DD419 pKa = 3.52KK420 pKa = 10.68SNKK423 pKa = 8.73GVSGGYY429 pKa = 10.08CPLTVSATPVVPNTNLAVATKK450 pKa = 8.93TVNGSLYY457 pKa = 9.96FPFYY461 pKa = 10.63TRR463 pKa = 11.84LSLTADD469 pKa = 4.01WSPGTATGTINRR481 pKa = 11.84WVVQNDD487 pKa = 3.09TSYY490 pKa = 11.99ALNNAYY496 pKa = 10.04GAGPKK501 pKa = 10.08GATNITFWKK510 pKa = 9.15CPRR513 pKa = 11.84DD514 pKa = 3.56AVVLFVFNIAFSVATSTNRR533 pKa = 11.84FFSIVITPVVPANTPPPYY551 pKa = 10.09QYY553 pKa = 11.6NFGLDD558 pKa = 3.77NYY560 pKa = 10.25ASSYY564 pKa = 11.27SFASSLIIPLRR575 pKa = 11.84QGDD578 pKa = 3.72QLQINWSMAGQSPAFQTTYY597 pKa = 10.23TYY599 pKa = 9.78WHH601 pKa = 6.7INEE604 pKa = 4.39MSFGVYY610 pKa = 10.13DD611 pKa = 3.98PNLL614 pKa = 3.53
MM1 pKa = 7.38QNTTNFFRR9 pKa = 11.84DD10 pKa = 3.14ISLVGSVTSLPPVLSRR26 pKa = 11.84GTLVYY31 pKa = 10.64PEE33 pKa = 5.22DD34 pKa = 3.8SSNLYY39 pKa = 9.91ISNGSQWVIAGGDD52 pKa = 3.71VGPLAAQVAQNTADD66 pKa = 3.3IGVLQGDD73 pKa = 3.92VSTLQGEE80 pKa = 4.8VATNTTDD87 pKa = 3.38IAGLQTDD94 pKa = 3.94VSTLQGQVSSLSTDD108 pKa = 3.05VSALQTQVATNTTDD122 pKa = 2.61IGTLQGDD129 pKa = 3.76VSTLQGEE136 pKa = 4.85VATNTADD143 pKa = 3.34ISTLQGDD150 pKa = 4.07VSTLQTQVNANTLDD164 pKa = 3.27ISALQTQVATNTTDD178 pKa = 2.61IGTLQGDD185 pKa = 3.76VSTLQGEE192 pKa = 4.8VATNTSNITTLQGQVATNTLDD213 pKa = 3.05IGTLQTDD220 pKa = 3.6VTTLQGDD227 pKa = 3.79VSTLQTQVGTNTSNITTLQGQVATNTTDD255 pKa = 2.86ISALQTQVSTNTTNIGTLQGDD276 pKa = 3.84VSTLQTQVTTNTLDD290 pKa = 2.96IGNLQTDD297 pKa = 4.08VSTLQTDD304 pKa = 4.24VAQNTLDD311 pKa = 3.23ISGLQGQVATNTTNIATNTSNISTLQGQTATNTTNITTLQGQVSTNTSDD360 pKa = 3.16ISTLQTQVSTNTADD374 pKa = 2.92IGTLQGQVATNTTNIATNTSNISALQGQVSTNTSNISTLQTTKK417 pKa = 10.58EE418 pKa = 4.04DD419 pKa = 3.52KK420 pKa = 10.68SNKK423 pKa = 8.73GVSGGYY429 pKa = 10.08CPLTVSATPVVPNTNLAVATKK450 pKa = 8.93TVNGSLYY457 pKa = 9.96FPFYY461 pKa = 10.63TRR463 pKa = 11.84LSLTADD469 pKa = 4.01WSPGTATGTINRR481 pKa = 11.84WVVQNDD487 pKa = 3.09TSYY490 pKa = 11.99ALNNAYY496 pKa = 10.04GAGPKK501 pKa = 10.08GATNITFWKK510 pKa = 9.15CPRR513 pKa = 11.84DD514 pKa = 3.56AVVLFVFNIAFSVATSTNRR533 pKa = 11.84FFSIVITPVVPANTPPPYY551 pKa = 10.09QYY553 pKa = 11.6NFGLDD558 pKa = 3.77NYY560 pKa = 10.25ASSYY564 pKa = 11.27SFASSLIIPLRR575 pKa = 11.84QGDD578 pKa = 3.72QLQINWSMAGQSPAFQTTYY597 pKa = 10.23TYY599 pKa = 9.78WHH601 pKa = 6.7INEE604 pKa = 4.39MSFGVYY610 pKa = 10.13DD611 pKa = 3.98PNLL614 pKa = 3.53
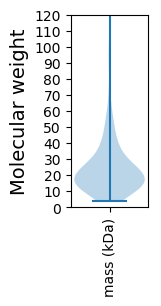
Molecular weight: 64.37 kDa
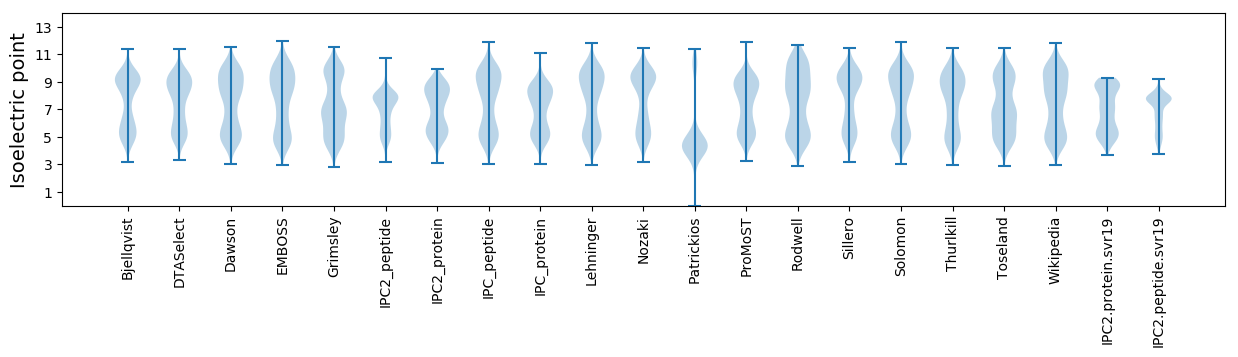
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A142CJG9|A0A142CJG9_9VIRU MORN repeat-containing protein OS=Brazilian marseillevirus OX=1813599 PE=4 SV=1
MM1 pKa = 6.85QARR4 pKa = 11.84IRR6 pKa = 11.84EE7 pKa = 4.12YY8 pKa = 10.32QRR10 pKa = 11.84LFPSVKK16 pKa = 9.53GLKK19 pKa = 9.29IVPSDD24 pKa = 3.74RR25 pKa = 11.84SQKK28 pKa = 9.79RR29 pKa = 11.84YY30 pKa = 9.81KK31 pKa = 9.93ATFVMDD37 pKa = 3.85GEE39 pKa = 4.5EE40 pKa = 4.19KK41 pKa = 10.51VVHH44 pKa = 6.29FGQRR48 pKa = 11.84GAFTFADD55 pKa = 4.51GAPEE59 pKa = 4.34SKK61 pKa = 10.11RR62 pKa = 11.84QAYY65 pKa = 8.43RR66 pKa = 11.84ARR68 pKa = 11.84HH69 pKa = 4.98SKK71 pKa = 9.98ILNKK75 pKa = 10.06GRR77 pKa = 11.84TAYY80 pKa = 9.31KK81 pKa = 10.41VPGSASSLAWVILWW95 pKa = 3.75
MM1 pKa = 6.85QARR4 pKa = 11.84IRR6 pKa = 11.84EE7 pKa = 4.12YY8 pKa = 10.32QRR10 pKa = 11.84LFPSVKK16 pKa = 9.53GLKK19 pKa = 9.29IVPSDD24 pKa = 3.74RR25 pKa = 11.84SQKK28 pKa = 9.79RR29 pKa = 11.84YY30 pKa = 9.81KK31 pKa = 9.93ATFVMDD37 pKa = 3.85GEE39 pKa = 4.5EE40 pKa = 4.19KK41 pKa = 10.51VVHH44 pKa = 6.29FGQRR48 pKa = 11.84GAFTFADD55 pKa = 4.51GAPEE59 pKa = 4.34SKK61 pKa = 10.11RR62 pKa = 11.84QAYY65 pKa = 8.43RR66 pKa = 11.84ARR68 pKa = 11.84HH69 pKa = 4.98SKK71 pKa = 9.98ILNKK75 pKa = 10.06GRR77 pKa = 11.84TAYY80 pKa = 9.31KK81 pKa = 10.41VPGSASSLAWVILWW95 pKa = 3.75
Molecular weight: 10.86 kDa
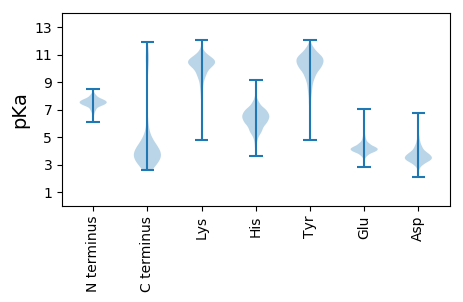
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
116783 |
34 |
1553 |
237.8 |
27.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.175 ± 0.119 | 2.435 ± 0.102 |
4.935 ± 0.066 | 8.161 ± 0.14 |
5.736 ± 0.087 | 6.122 ± 0.113 |
1.848 ± 0.064 | 5.004 ± 0.072 |
8.541 ± 0.18 | 8.905 ± 0.111 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.16 ± 0.053 | 4.203 ± 0.081 |
3.847 ± 0.087 | 3.364 ± 0.082 |
5.435 ± 0.093 | 7.615 ± 0.103 |
5.24 ± 0.118 | 6.673 ± 0.074 |
1.463 ± 0.048 | 3.139 ± 0.061 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
