
Pleionea mediterranea
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Pleioneaceae; Pleionea
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
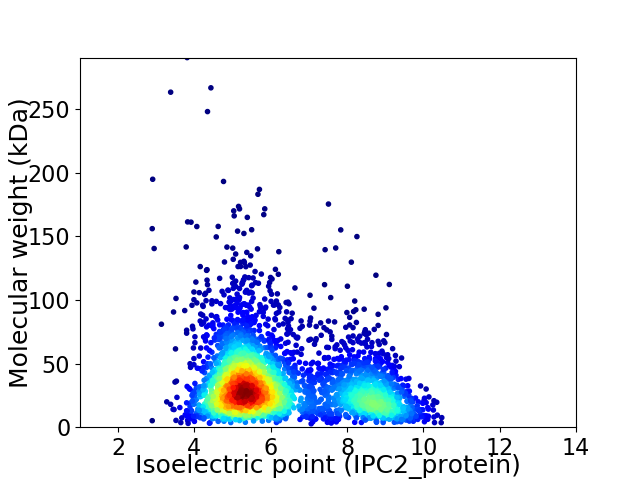
Virtual 2D-PAGE plot for 3566 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A316G0R5|A0A316G0R5_9GAMM UDP-N-acetylmuramoylalanine--D-glutamate ligase OS=Pleionea mediterranea OX=523701 GN=murD PE=3 SV=1
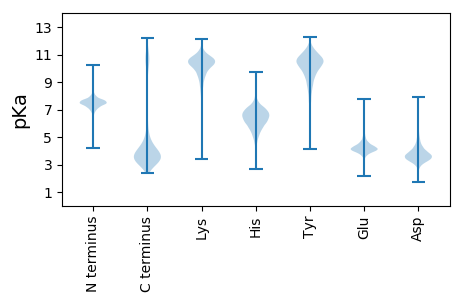
MM1 pKa = 6.82MRR3 pKa = 11.84WSIILKK9 pKa = 10.07SALILLLSIYY19 pKa = 10.93SMAAQSVIIFSDD31 pKa = 3.63DD32 pKa = 3.5FEE34 pKa = 4.67TLLTQWTVNSSGGDD48 pKa = 3.21ASIGNEE54 pKa = 3.86TSNSGNNSLRR64 pKa = 11.84LRR66 pKa = 11.84WDD68 pKa = 3.44TVTITSNTINANVSAAQVRR87 pKa = 11.84IWVRR91 pKa = 11.84RR92 pKa = 11.84GDD94 pKa = 3.96DD95 pKa = 3.46NFSEE99 pKa = 4.25NPEE102 pKa = 3.99NGEE105 pKa = 4.01NLTLQYY111 pKa = 11.33LNNSNNWITLDD122 pKa = 3.5TFFGGGSPGDD132 pKa = 3.7IFDD135 pKa = 5.17LGYY138 pKa = 11.02NLPADD143 pKa = 4.23ALHH146 pKa = 6.37SGLRR150 pKa = 11.84LRR152 pKa = 11.84LNLNTGSGNDD162 pKa = 3.14FDD164 pKa = 4.37YY165 pKa = 10.82WHH167 pKa = 6.96VDD169 pKa = 3.3DD170 pKa = 5.61VVVDD174 pKa = 4.91SIDD177 pKa = 3.31ATAPTGLVAEE187 pKa = 4.32WRR189 pKa = 11.84FDD191 pKa = 3.49EE192 pKa = 4.82LTWNSQALEE201 pKa = 4.12VRR203 pKa = 11.84DD204 pKa = 3.74SSGNFYY210 pKa = 10.74HH211 pKa = 7.41GIAFNSQPTTGFVCNAADD229 pKa = 4.06FTAAGTDD236 pKa = 3.32DD237 pKa = 4.59YY238 pKa = 11.67IRR240 pKa = 11.84LNHH243 pKa = 6.48EE244 pKa = 4.43SLNGLDD250 pKa = 4.04DD251 pKa = 3.62WSISVWVTSNRR262 pKa = 11.84NAAQTILSASKK273 pKa = 11.13NEE275 pKa = 4.15TEE277 pKa = 4.09LNEE280 pKa = 3.64AVMYY284 pKa = 10.83FNDD287 pKa = 5.22ASTFWPSVRR296 pKa = 11.84SNPFFDD302 pKa = 5.37NNTQLALPYY311 pKa = 10.48AIDD314 pKa = 4.78DD315 pKa = 4.39GNFHH319 pKa = 6.53HH320 pKa = 7.01LVWTRR325 pKa = 11.84DD326 pKa = 3.27RR327 pKa = 11.84ANSEE331 pKa = 3.79TCLYY335 pKa = 10.34LDD337 pKa = 4.28SNIVGCTTHH346 pKa = 7.04TDD348 pKa = 3.13GGNTMDD354 pKa = 4.45VIVDD358 pKa = 3.59GLVIGQEE365 pKa = 3.82QDD367 pKa = 3.45VVGGDD372 pKa = 3.5FDD374 pKa = 5.44INQVWVGLIDD384 pKa = 4.0EE385 pKa = 5.27LVLFDD390 pKa = 5.08NEE392 pKa = 4.16LSPGEE397 pKa = 4.0VLFIYY402 pKa = 10.58NNQTAGRR409 pKa = 11.84GWDD412 pKa = 3.47GSDD415 pKa = 3.31RR416 pKa = 11.84VCPEE420 pKa = 4.11ALPPTAAYY428 pKa = 9.22WRR430 pKa = 11.84MDD432 pKa = 3.09EE433 pKa = 4.43TTWDD437 pKa = 3.71GSPGEE442 pKa = 4.43VIDD445 pKa = 5.61LSGNGLNMTSYY456 pKa = 11.29GGASTNNIDD465 pKa = 3.63PAIVGDD471 pKa = 4.24PGTCGFGEE479 pKa = 4.32FDD481 pKa = 3.7GSTTYY486 pKa = 11.18LEE488 pKa = 4.57IDD490 pKa = 3.94HH491 pKa = 7.46DD492 pKa = 4.5PLLSFSTNMSITLWVKK508 pKa = 10.39PNSLPLSGLHH518 pKa = 6.36SVLSKK523 pKa = 11.04DD524 pKa = 3.47EE525 pKa = 4.3NYY527 pKa = 10.33EE528 pKa = 3.94FHH530 pKa = 6.8LTPTNEE536 pKa = 3.92IFWWWNQGALTTSGANIQPGNWYY559 pKa = 10.12HH560 pKa = 6.81IGLTFRR566 pKa = 11.84SGEE569 pKa = 3.8QIIYY573 pKa = 10.4VNGIEE578 pKa = 5.25RR579 pKa = 11.84GRR581 pKa = 11.84QNFNGTLPQNTDD593 pKa = 3.39PLQIGQDD600 pKa = 3.32QLAAGRR606 pKa = 11.84FLDD609 pKa = 4.18GQVDD613 pKa = 3.94EE614 pKa = 5.31VKK616 pKa = 10.35IFPSTLSHH624 pKa = 5.77GQVIQSMLEE633 pKa = 3.79VHH635 pKa = 7.04PCTSSSTCPYY645 pKa = 10.59DD646 pKa = 3.39YY647 pKa = 10.83TDD649 pKa = 3.49NFSVQSYY656 pKa = 10.15GNNDD660 pKa = 2.88GSNNFSGNWQEE671 pKa = 4.31TGDD674 pKa = 4.02NNSPASGSVGIAGGEE689 pKa = 3.99LYY691 pKa = 9.46LTNQNNNEE699 pKa = 3.94PSISRR704 pKa = 11.84SMDD707 pKa = 3.36LSQALTAEE715 pKa = 4.57LRR717 pKa = 11.84FEE719 pKa = 4.1MRR721 pKa = 11.84TSNTLEE727 pKa = 3.96NGDD730 pKa = 3.42RR731 pKa = 11.84VAIEE735 pKa = 3.95VSADD739 pKa = 3.08GTNYY743 pKa = 10.33TEE745 pKa = 4.15LQEE748 pKa = 4.16FRR750 pKa = 11.84NDD752 pKa = 3.12VVGVFTYY759 pKa = 10.62DD760 pKa = 2.74IAGFISANTSIRR772 pKa = 11.84FRR774 pKa = 11.84ITQGYY779 pKa = 8.01RR780 pKa = 11.84QNDD783 pKa = 3.74EE784 pKa = 3.96YY785 pKa = 11.7VYY787 pKa = 10.95FDD789 pKa = 4.14DD790 pKa = 3.9VQVNITEE797 pKa = 4.14FCGIGFFVINHH808 pKa = 6.98DD809 pKa = 3.75GSGINCLRR817 pKa = 11.84EE818 pKa = 4.08AVSITAYY825 pKa = 9.36DD826 pKa = 3.21TGGNLLTGYY835 pKa = 10.15NGLVSLSSNTSNGNWFNTDD854 pKa = 2.59SSGNSADD861 pKa = 4.28LSQGTLNDD869 pKa = 3.51TAGDD873 pKa = 3.97DD874 pKa = 3.89NGAATYY880 pKa = 10.07QFSPADD886 pKa = 3.38QGTVTLYY893 pKa = 11.16LEE895 pKa = 4.45NTHH898 pKa = 6.09QEE900 pKa = 4.44SINVAVTEE908 pKa = 4.3GAAADD913 pKa = 4.48DD914 pKa = 3.89NSEE917 pKa = 3.99GNIVFRR923 pKa = 11.84PFGFVATPSSIPTQVAGRR941 pKa = 11.84PFNLQLAAAGQTPSQPEE958 pKa = 3.92CGVIEE963 pKa = 4.81EE964 pKa = 4.34YY965 pKa = 10.32TGNKK969 pKa = 9.43DD970 pKa = 2.79IHH972 pKa = 6.11FWSSYY977 pKa = 6.98TLPNSSPTPVTINGVNVAGSEE998 pKa = 4.23PAASSQPITFNNGIADD1014 pKa = 3.64IPVQYY1019 pKa = 10.97NDD1021 pKa = 2.86VGQIQFFAKK1030 pKa = 10.43DD1031 pKa = 3.31EE1032 pKa = 4.27TGIGDD1037 pKa = 3.98PTTGNTDD1044 pKa = 3.76EE1045 pKa = 5.31IIGGIPPFVVRR1056 pKa = 11.84PFGYY1060 pKa = 10.41DD1061 pKa = 2.52ISAQGNPDD1069 pKa = 3.2ATGGASPVYY1078 pKa = 10.08AVAGNAFNMTLRR1090 pKa = 11.84SVLWQAADD1098 pKa = 3.31SDD1100 pKa = 4.8ANGVPLPGADD1110 pKa = 3.76LSDD1113 pKa = 3.73NGVTPNIVNIPSSAPDD1129 pKa = 3.13IDD1131 pKa = 3.73ITLTPTAQVVTNSDD1145 pKa = 3.5GTLSDD1150 pKa = 3.3PTINYY1155 pKa = 10.26SNFSAVGSAAQGTLTQSQSWSEE1177 pKa = 4.01VGILTLNANTPDD1189 pKa = 3.7FMATGVGVTGSKK1201 pKa = 10.8NNIGRR1206 pKa = 11.84FIPSYY1211 pKa = 10.69FSVQVGDD1218 pKa = 4.05NFEE1221 pKa = 4.21AQCTGFSYY1229 pKa = 10.85SGFNNGSAGLTKK1241 pKa = 9.98IGQSDD1246 pKa = 4.09NLTIDD1251 pKa = 3.15IQAFNANDD1259 pKa = 3.78EE1260 pKa = 4.41TTQNYY1265 pKa = 9.57DD1266 pKa = 2.71ADD1268 pKa = 4.11FAKK1271 pKa = 10.74LEE1273 pKa = 4.35SGTISTSGYY1282 pKa = 10.63DD1283 pKa = 3.5EE1284 pKa = 4.5TNAAPAPGNIVTGTVGTPVFDD1305 pKa = 3.76TDD1307 pKa = 4.13GFAQNVSIPNVYY1319 pKa = 9.37YY1320 pKa = 10.68QYY1322 pKa = 11.67DD1323 pKa = 3.47NFAAPFEE1330 pKa = 4.03LRR1332 pKa = 11.84IDD1334 pKa = 3.63IDD1336 pKa = 3.59ATDD1339 pKa = 3.29VDD1341 pKa = 4.83GVNGTASSNSVEE1353 pKa = 3.98QRR1355 pKa = 11.84LGRR1358 pKa = 11.84SRR1360 pKa = 11.84LVTAYY1365 pKa = 10.0GPEE1368 pKa = 4.05VEE1370 pKa = 4.82PLNIPLYY1377 pKa = 10.15TEE1379 pKa = 4.68FFNGTDD1385 pKa = 3.27WVINTADD1392 pKa = 3.37NCTSYY1397 pKa = 10.72ISSDD1401 pKa = 3.48LQFVTGTYY1409 pKa = 10.12QGDD1412 pKa = 3.87LNAGEE1417 pKa = 4.26TSITLPASNTLLTAGEE1433 pKa = 4.3SGVNQGFGLTAPGVGNSGEE1452 pKa = 4.28VNVEE1456 pKa = 4.17HH1457 pKa = 6.79NLSALDD1463 pKa = 3.44WLRR1466 pKa = 11.84FDD1468 pKa = 4.14WNGDD1472 pKa = 3.3NSNDD1476 pKa = 3.69NPQATAGFGRR1486 pKa = 11.84YY1487 pKa = 9.0RR1488 pKa = 11.84GSDD1491 pKa = 2.88RR1492 pKa = 11.84VIYY1495 pKa = 9.6WRR1497 pKa = 11.84EE1498 pKa = 3.47NN1499 pKa = 3.09
MM1 pKa = 6.82MRR3 pKa = 11.84WSIILKK9 pKa = 10.07SALILLLSIYY19 pKa = 10.93SMAAQSVIIFSDD31 pKa = 3.63DD32 pKa = 3.5FEE34 pKa = 4.67TLLTQWTVNSSGGDD48 pKa = 3.21ASIGNEE54 pKa = 3.86TSNSGNNSLRR64 pKa = 11.84LRR66 pKa = 11.84WDD68 pKa = 3.44TVTITSNTINANVSAAQVRR87 pKa = 11.84IWVRR91 pKa = 11.84RR92 pKa = 11.84GDD94 pKa = 3.96DD95 pKa = 3.46NFSEE99 pKa = 4.25NPEE102 pKa = 3.99NGEE105 pKa = 4.01NLTLQYY111 pKa = 11.33LNNSNNWITLDD122 pKa = 3.5TFFGGGSPGDD132 pKa = 3.7IFDD135 pKa = 5.17LGYY138 pKa = 11.02NLPADD143 pKa = 4.23ALHH146 pKa = 6.37SGLRR150 pKa = 11.84LRR152 pKa = 11.84LNLNTGSGNDD162 pKa = 3.14FDD164 pKa = 4.37YY165 pKa = 10.82WHH167 pKa = 6.96VDD169 pKa = 3.3DD170 pKa = 5.61VVVDD174 pKa = 4.91SIDD177 pKa = 3.31ATAPTGLVAEE187 pKa = 4.32WRR189 pKa = 11.84FDD191 pKa = 3.49EE192 pKa = 4.82LTWNSQALEE201 pKa = 4.12VRR203 pKa = 11.84DD204 pKa = 3.74SSGNFYY210 pKa = 10.74HH211 pKa = 7.41GIAFNSQPTTGFVCNAADD229 pKa = 4.06FTAAGTDD236 pKa = 3.32DD237 pKa = 4.59YY238 pKa = 11.67IRR240 pKa = 11.84LNHH243 pKa = 6.48EE244 pKa = 4.43SLNGLDD250 pKa = 4.04DD251 pKa = 3.62WSISVWVTSNRR262 pKa = 11.84NAAQTILSASKK273 pKa = 11.13NEE275 pKa = 4.15TEE277 pKa = 4.09LNEE280 pKa = 3.64AVMYY284 pKa = 10.83FNDD287 pKa = 5.22ASTFWPSVRR296 pKa = 11.84SNPFFDD302 pKa = 5.37NNTQLALPYY311 pKa = 10.48AIDD314 pKa = 4.78DD315 pKa = 4.39GNFHH319 pKa = 6.53HH320 pKa = 7.01LVWTRR325 pKa = 11.84DD326 pKa = 3.27RR327 pKa = 11.84ANSEE331 pKa = 3.79TCLYY335 pKa = 10.34LDD337 pKa = 4.28SNIVGCTTHH346 pKa = 7.04TDD348 pKa = 3.13GGNTMDD354 pKa = 4.45VIVDD358 pKa = 3.59GLVIGQEE365 pKa = 3.82QDD367 pKa = 3.45VVGGDD372 pKa = 3.5FDD374 pKa = 5.44INQVWVGLIDD384 pKa = 4.0EE385 pKa = 5.27LVLFDD390 pKa = 5.08NEE392 pKa = 4.16LSPGEE397 pKa = 4.0VLFIYY402 pKa = 10.58NNQTAGRR409 pKa = 11.84GWDD412 pKa = 3.47GSDD415 pKa = 3.31RR416 pKa = 11.84VCPEE420 pKa = 4.11ALPPTAAYY428 pKa = 9.22WRR430 pKa = 11.84MDD432 pKa = 3.09EE433 pKa = 4.43TTWDD437 pKa = 3.71GSPGEE442 pKa = 4.43VIDD445 pKa = 5.61LSGNGLNMTSYY456 pKa = 11.29GGASTNNIDD465 pKa = 3.63PAIVGDD471 pKa = 4.24PGTCGFGEE479 pKa = 4.32FDD481 pKa = 3.7GSTTYY486 pKa = 11.18LEE488 pKa = 4.57IDD490 pKa = 3.94HH491 pKa = 7.46DD492 pKa = 4.5PLLSFSTNMSITLWVKK508 pKa = 10.39PNSLPLSGLHH518 pKa = 6.36SVLSKK523 pKa = 11.04DD524 pKa = 3.47EE525 pKa = 4.3NYY527 pKa = 10.33EE528 pKa = 3.94FHH530 pKa = 6.8LTPTNEE536 pKa = 3.92IFWWWNQGALTTSGANIQPGNWYY559 pKa = 10.12HH560 pKa = 6.81IGLTFRR566 pKa = 11.84SGEE569 pKa = 3.8QIIYY573 pKa = 10.4VNGIEE578 pKa = 5.25RR579 pKa = 11.84GRR581 pKa = 11.84QNFNGTLPQNTDD593 pKa = 3.39PLQIGQDD600 pKa = 3.32QLAAGRR606 pKa = 11.84FLDD609 pKa = 4.18GQVDD613 pKa = 3.94EE614 pKa = 5.31VKK616 pKa = 10.35IFPSTLSHH624 pKa = 5.77GQVIQSMLEE633 pKa = 3.79VHH635 pKa = 7.04PCTSSSTCPYY645 pKa = 10.59DD646 pKa = 3.39YY647 pKa = 10.83TDD649 pKa = 3.49NFSVQSYY656 pKa = 10.15GNNDD660 pKa = 2.88GSNNFSGNWQEE671 pKa = 4.31TGDD674 pKa = 4.02NNSPASGSVGIAGGEE689 pKa = 3.99LYY691 pKa = 9.46LTNQNNNEE699 pKa = 3.94PSISRR704 pKa = 11.84SMDD707 pKa = 3.36LSQALTAEE715 pKa = 4.57LRR717 pKa = 11.84FEE719 pKa = 4.1MRR721 pKa = 11.84TSNTLEE727 pKa = 3.96NGDD730 pKa = 3.42RR731 pKa = 11.84VAIEE735 pKa = 3.95VSADD739 pKa = 3.08GTNYY743 pKa = 10.33TEE745 pKa = 4.15LQEE748 pKa = 4.16FRR750 pKa = 11.84NDD752 pKa = 3.12VVGVFTYY759 pKa = 10.62DD760 pKa = 2.74IAGFISANTSIRR772 pKa = 11.84FRR774 pKa = 11.84ITQGYY779 pKa = 8.01RR780 pKa = 11.84QNDD783 pKa = 3.74EE784 pKa = 3.96YY785 pKa = 11.7VYY787 pKa = 10.95FDD789 pKa = 4.14DD790 pKa = 3.9VQVNITEE797 pKa = 4.14FCGIGFFVINHH808 pKa = 6.98DD809 pKa = 3.75GSGINCLRR817 pKa = 11.84EE818 pKa = 4.08AVSITAYY825 pKa = 9.36DD826 pKa = 3.21TGGNLLTGYY835 pKa = 10.15NGLVSLSSNTSNGNWFNTDD854 pKa = 2.59SSGNSADD861 pKa = 4.28LSQGTLNDD869 pKa = 3.51TAGDD873 pKa = 3.97DD874 pKa = 3.89NGAATYY880 pKa = 10.07QFSPADD886 pKa = 3.38QGTVTLYY893 pKa = 11.16LEE895 pKa = 4.45NTHH898 pKa = 6.09QEE900 pKa = 4.44SINVAVTEE908 pKa = 4.3GAAADD913 pKa = 4.48DD914 pKa = 3.89NSEE917 pKa = 3.99GNIVFRR923 pKa = 11.84PFGFVATPSSIPTQVAGRR941 pKa = 11.84PFNLQLAAAGQTPSQPEE958 pKa = 3.92CGVIEE963 pKa = 4.81EE964 pKa = 4.34YY965 pKa = 10.32TGNKK969 pKa = 9.43DD970 pKa = 2.79IHH972 pKa = 6.11FWSSYY977 pKa = 6.98TLPNSSPTPVTINGVNVAGSEE998 pKa = 4.23PAASSQPITFNNGIADD1014 pKa = 3.64IPVQYY1019 pKa = 10.97NDD1021 pKa = 2.86VGQIQFFAKK1030 pKa = 10.43DD1031 pKa = 3.31EE1032 pKa = 4.27TGIGDD1037 pKa = 3.98PTTGNTDD1044 pKa = 3.76EE1045 pKa = 5.31IIGGIPPFVVRR1056 pKa = 11.84PFGYY1060 pKa = 10.41DD1061 pKa = 2.52ISAQGNPDD1069 pKa = 3.2ATGGASPVYY1078 pKa = 10.08AVAGNAFNMTLRR1090 pKa = 11.84SVLWQAADD1098 pKa = 3.31SDD1100 pKa = 4.8ANGVPLPGADD1110 pKa = 3.76LSDD1113 pKa = 3.73NGVTPNIVNIPSSAPDD1129 pKa = 3.13IDD1131 pKa = 3.73ITLTPTAQVVTNSDD1145 pKa = 3.5GTLSDD1150 pKa = 3.3PTINYY1155 pKa = 10.26SNFSAVGSAAQGTLTQSQSWSEE1177 pKa = 4.01VGILTLNANTPDD1189 pKa = 3.7FMATGVGVTGSKK1201 pKa = 10.8NNIGRR1206 pKa = 11.84FIPSYY1211 pKa = 10.69FSVQVGDD1218 pKa = 4.05NFEE1221 pKa = 4.21AQCTGFSYY1229 pKa = 10.85SGFNNGSAGLTKK1241 pKa = 9.98IGQSDD1246 pKa = 4.09NLTIDD1251 pKa = 3.15IQAFNANDD1259 pKa = 3.78EE1260 pKa = 4.41TTQNYY1265 pKa = 9.57DD1266 pKa = 2.71ADD1268 pKa = 4.11FAKK1271 pKa = 10.74LEE1273 pKa = 4.35SGTISTSGYY1282 pKa = 10.63DD1283 pKa = 3.5EE1284 pKa = 4.5TNAAPAPGNIVTGTVGTPVFDD1305 pKa = 3.76TDD1307 pKa = 4.13GFAQNVSIPNVYY1319 pKa = 9.37YY1320 pKa = 10.68QYY1322 pKa = 11.67DD1323 pKa = 3.47NFAAPFEE1330 pKa = 4.03LRR1332 pKa = 11.84IDD1334 pKa = 3.63IDD1336 pKa = 3.59ATDD1339 pKa = 3.29VDD1341 pKa = 4.83GVNGTASSNSVEE1353 pKa = 3.98QRR1355 pKa = 11.84LGRR1358 pKa = 11.84SRR1360 pKa = 11.84LVTAYY1365 pKa = 10.0GPEE1368 pKa = 4.05VEE1370 pKa = 4.82PLNIPLYY1377 pKa = 10.15TEE1379 pKa = 4.68FFNGTDD1385 pKa = 3.27WVINTADD1392 pKa = 3.37NCTSYY1397 pKa = 10.72ISSDD1401 pKa = 3.48LQFVTGTYY1409 pKa = 10.12QGDD1412 pKa = 3.87LNAGEE1417 pKa = 4.26TSITLPASNTLLTAGEE1433 pKa = 4.3SGVNQGFGLTAPGVGNSGEE1452 pKa = 4.28VNVEE1456 pKa = 4.17HH1457 pKa = 6.79NLSALDD1463 pKa = 3.44WLRR1466 pKa = 11.84FDD1468 pKa = 4.14WNGDD1472 pKa = 3.3NSNDD1476 pKa = 3.69NPQATAGFGRR1486 pKa = 11.84YY1487 pKa = 9.0RR1488 pKa = 11.84GSDD1491 pKa = 2.88RR1492 pKa = 11.84VIYY1495 pKa = 9.6WRR1497 pKa = 11.84EE1498 pKa = 3.47NN1499 pKa = 3.09
Molecular weight: 161.47 kDa
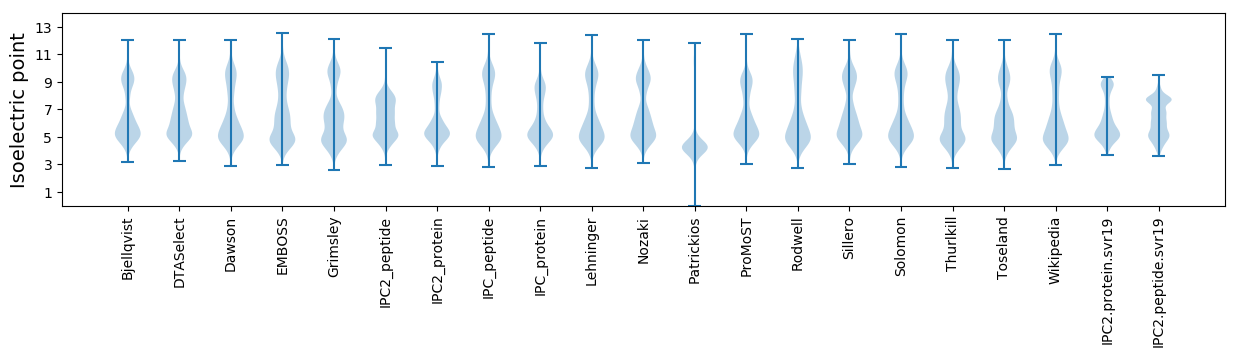
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A316FRC0|A0A316FRC0_9GAMM Phosphoserine aminotransferase OS=Pleionea mediterranea OX=523701 GN=serC PE=3 SV=1
MM1 pKa = 7.93ISITSLYY8 pKa = 10.97ASLCGILILALAIRR22 pKa = 11.84VVYY25 pKa = 10.3LRR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 8.86NKK31 pKa = 9.94VGIGNGGVDD40 pKa = 3.78DD41 pKa = 4.21LRR43 pKa = 11.84RR44 pKa = 11.84AVRR47 pKa = 11.84VHH49 pKa = 6.59ANAIEE54 pKa = 4.19YY55 pKa = 10.25IPIVLILMAALEE67 pKa = 4.35LNGVNGWLMHH77 pKa = 5.51VLGSLLVLSRR87 pKa = 11.84LLHH90 pKa = 5.47AQGLSGSPGVSFGRR104 pKa = 11.84FWGTLVTWIVIIISVVANLVQFFANN129 pKa = 3.52
MM1 pKa = 7.93ISITSLYY8 pKa = 10.97ASLCGILILALAIRR22 pKa = 11.84VVYY25 pKa = 10.3LRR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 8.86NKK31 pKa = 9.94VGIGNGGVDD40 pKa = 3.78DD41 pKa = 4.21LRR43 pKa = 11.84RR44 pKa = 11.84AVRR47 pKa = 11.84VHH49 pKa = 6.59ANAIEE54 pKa = 4.19YY55 pKa = 10.25IPIVLILMAALEE67 pKa = 4.35LNGVNGWLMHH77 pKa = 5.51VLGSLLVLSRR87 pKa = 11.84LLHH90 pKa = 5.47AQGLSGSPGVSFGRR104 pKa = 11.84FWGTLVTWIVIIISVVANLVQFFANN129 pKa = 3.52
Molecular weight: 13.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1176721 |
25 |
2691 |
330.0 |
36.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.819 ± 0.046 | 0.938 ± 0.013 |
5.912 ± 0.053 | 6.201 ± 0.047 |
4.256 ± 0.031 | 6.361 ± 0.046 |
2.227 ± 0.021 | 6.49 ± 0.033 |
5.793 ± 0.046 | 10.146 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.355 ± 0.026 | 4.852 ± 0.038 |
3.782 ± 0.023 | 4.958 ± 0.04 |
4.397 ± 0.031 | 7.32 ± 0.042 |
5.29 ± 0.033 | 6.519 ± 0.038 |
1.259 ± 0.016 | 3.125 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
