
Blastomyces percursus
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Onygenales;
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
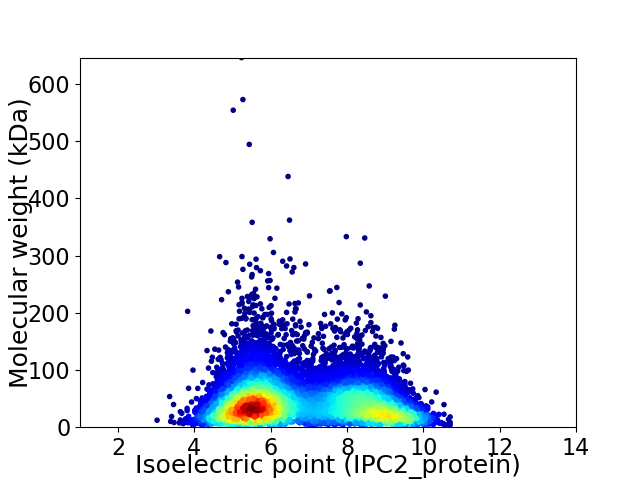
Virtual 2D-PAGE plot for 10283 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1J9R7U1|A0A1J9R7U1_9EURO tRNA pseudouridine(38-40) synthase OS=Blastomyces percursus OX=1658174 GN=ACJ73_04574 PE=3 SV=1
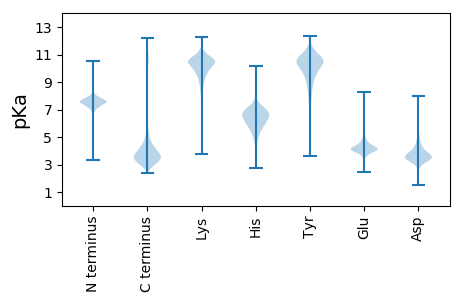
MM1 pKa = 7.79DD2 pKa = 3.82MAAIEE7 pKa = 4.27MMDD10 pKa = 2.87IGYY13 pKa = 10.17DD14 pKa = 3.23EE15 pKa = 5.59GGDD18 pKa = 3.63YY19 pKa = 10.9SYY21 pKa = 11.52DD22 pKa = 3.43EE23 pKa = 5.37PYY25 pKa = 10.93NDD27 pKa = 5.19SEE29 pKa = 4.78SCDD32 pKa = 3.6TEE34 pKa = 5.61DD35 pKa = 3.94YY36 pKa = 10.56TGSSYY41 pKa = 11.31NRR43 pKa = 11.84NNSPSYY49 pKa = 11.11DD50 pKa = 3.6YY51 pKa = 10.19GTDD54 pKa = 3.52GASEE58 pKa = 4.49GNGPRR63 pKa = 11.84DD64 pKa = 3.38AEE66 pKa = 4.08EE67 pKa = 4.18YY68 pKa = 9.43PRR70 pKa = 11.84KK71 pKa = 9.58EE72 pKa = 4.11LEE74 pKa = 3.97VDD76 pKa = 3.22SDD78 pKa = 3.67EE79 pKa = 4.36ATYY82 pKa = 11.17SSGSSCDD89 pKa = 3.34GSLQAEE95 pKa = 4.26AEE97 pKa = 4.07SSYY100 pKa = 11.18RR101 pKa = 11.84HH102 pKa = 5.97ASVPSGSPYY111 pKa = 10.55RR112 pKa = 11.84STRR115 pKa = 11.84WGGG118 pKa = 3.05
MM1 pKa = 7.79DD2 pKa = 3.82MAAIEE7 pKa = 4.27MMDD10 pKa = 2.87IGYY13 pKa = 10.17DD14 pKa = 3.23EE15 pKa = 5.59GGDD18 pKa = 3.63YY19 pKa = 10.9SYY21 pKa = 11.52DD22 pKa = 3.43EE23 pKa = 5.37PYY25 pKa = 10.93NDD27 pKa = 5.19SEE29 pKa = 4.78SCDD32 pKa = 3.6TEE34 pKa = 5.61DD35 pKa = 3.94YY36 pKa = 10.56TGSSYY41 pKa = 11.31NRR43 pKa = 11.84NNSPSYY49 pKa = 11.11DD50 pKa = 3.6YY51 pKa = 10.19GTDD54 pKa = 3.52GASEE58 pKa = 4.49GNGPRR63 pKa = 11.84DD64 pKa = 3.38AEE66 pKa = 4.08EE67 pKa = 4.18YY68 pKa = 9.43PRR70 pKa = 11.84KK71 pKa = 9.58EE72 pKa = 4.11LEE74 pKa = 3.97VDD76 pKa = 3.22SDD78 pKa = 3.67EE79 pKa = 4.36ATYY82 pKa = 11.17SSGSSCDD89 pKa = 3.34GSLQAEE95 pKa = 4.26AEE97 pKa = 4.07SSYY100 pKa = 11.18RR101 pKa = 11.84HH102 pKa = 5.97ASVPSGSPYY111 pKa = 10.55RR112 pKa = 11.84STRR115 pKa = 11.84WGGG118 pKa = 3.05
Molecular weight: 12.88 kDa
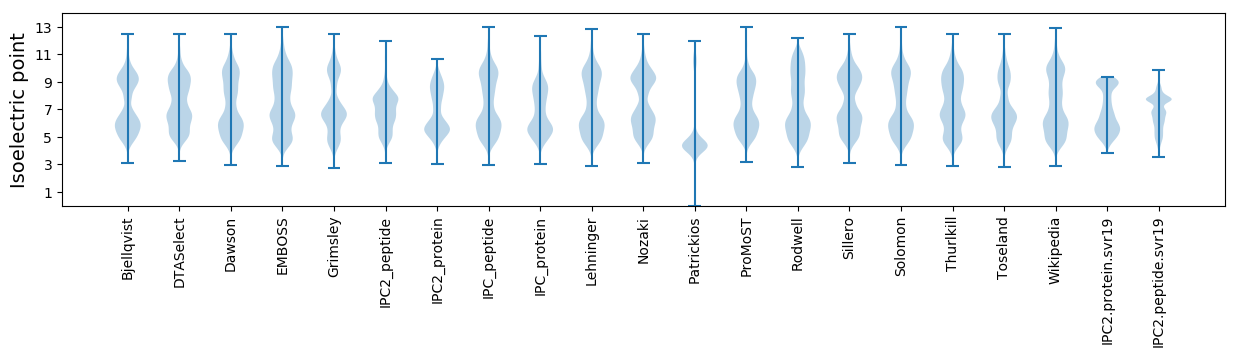
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1J9Q1C8|A0A1J9Q1C8_9EURO Uncharacterized protein OS=Blastomyces percursus OX=1658174 GN=ACJ73_09958 PE=4 SV=1
MM1 pKa = 7.14VFQWLQRR8 pKa = 11.84AWFTRR13 pKa = 11.84QGITYY18 pKa = 9.23FGLPIHH24 pKa = 6.77PMLGNGGNTSSTRR37 pKa = 11.84LIQFSISASTGRR49 pKa = 11.84YY50 pKa = 6.8VVCSTTHH57 pKa = 6.89PSSQTAPLMLANQRR71 pKa = 11.84IAWEE75 pKa = 4.03MSEE78 pKa = 3.93SGEE81 pKa = 4.01IFVRR85 pKa = 11.84SCADD89 pKa = 3.27VKK91 pKa = 10.81RR92 pKa = 11.84PPHH95 pKa = 6.51LLINNQLVIKK105 pKa = 10.32VKK107 pKa = 10.71LHH109 pKa = 6.06DD110 pKa = 3.83TFEE113 pKa = 4.19NAAFKK118 pKa = 11.12LNFSICTTRR127 pKa = 11.84RR128 pKa = 11.84YY129 pKa = 9.7HH130 pKa = 6.92DD131 pKa = 3.98CFRR134 pKa = 11.84AIWLRR139 pKa = 11.84APDD142 pKa = 4.05APAATTPQPVSEE154 pKa = 4.35TNSNIRR160 pKa = 11.84SRR162 pKa = 11.84LRR164 pKa = 11.84LFVRR168 pKa = 11.84QSHH171 pKa = 5.52GVNPKK176 pKa = 9.42PPIRR180 pKa = 11.84SRR182 pKa = 11.84SNAGCCLEE190 pKa = 3.96RR191 pKa = 11.84SGTGVVNIDD200 pKa = 3.45VGSNPTNRR208 pKa = 11.84RR209 pKa = 11.84GKK211 pKa = 9.25VLSGSTCASPFPVGVDD227 pKa = 3.29PDD229 pKa = 3.75LANAPEE235 pKa = 4.48HH236 pKa = 6.3PRR238 pKa = 11.84SAASRR243 pKa = 11.84EE244 pKa = 4.14QSSGTTLMRR253 pKa = 11.84PPQILASSDD262 pKa = 3.21APRR265 pKa = 11.84DD266 pKa = 3.61VPATFFIFSDD276 pKa = 3.65LCVRR280 pKa = 11.84TAGWYY285 pKa = 9.49RR286 pKa = 11.84LRR288 pKa = 11.84FQLVDD293 pKa = 3.4VQNRR297 pKa = 11.84GSPHH301 pKa = 6.52AFTRR305 pKa = 11.84SGLNLSKK312 pKa = 10.88CSLRR316 pKa = 11.84KK317 pKa = 9.66IFLGG321 pKa = 3.78
MM1 pKa = 7.14VFQWLQRR8 pKa = 11.84AWFTRR13 pKa = 11.84QGITYY18 pKa = 9.23FGLPIHH24 pKa = 6.77PMLGNGGNTSSTRR37 pKa = 11.84LIQFSISASTGRR49 pKa = 11.84YY50 pKa = 6.8VVCSTTHH57 pKa = 6.89PSSQTAPLMLANQRR71 pKa = 11.84IAWEE75 pKa = 4.03MSEE78 pKa = 3.93SGEE81 pKa = 4.01IFVRR85 pKa = 11.84SCADD89 pKa = 3.27VKK91 pKa = 10.81RR92 pKa = 11.84PPHH95 pKa = 6.51LLINNQLVIKK105 pKa = 10.32VKK107 pKa = 10.71LHH109 pKa = 6.06DD110 pKa = 3.83TFEE113 pKa = 4.19NAAFKK118 pKa = 11.12LNFSICTTRR127 pKa = 11.84RR128 pKa = 11.84YY129 pKa = 9.7HH130 pKa = 6.92DD131 pKa = 3.98CFRR134 pKa = 11.84AIWLRR139 pKa = 11.84APDD142 pKa = 4.05APAATTPQPVSEE154 pKa = 4.35TNSNIRR160 pKa = 11.84SRR162 pKa = 11.84LRR164 pKa = 11.84LFVRR168 pKa = 11.84QSHH171 pKa = 5.52GVNPKK176 pKa = 9.42PPIRR180 pKa = 11.84SRR182 pKa = 11.84SNAGCCLEE190 pKa = 3.96RR191 pKa = 11.84SGTGVVNIDD200 pKa = 3.45VGSNPTNRR208 pKa = 11.84RR209 pKa = 11.84GKK211 pKa = 9.25VLSGSTCASPFPVGVDD227 pKa = 3.29PDD229 pKa = 3.75LANAPEE235 pKa = 4.48HH236 pKa = 6.3PRR238 pKa = 11.84SAASRR243 pKa = 11.84EE244 pKa = 4.14QSSGTTLMRR253 pKa = 11.84PPQILASSDD262 pKa = 3.21APRR265 pKa = 11.84DD266 pKa = 3.61VPATFFIFSDD276 pKa = 3.65LCVRR280 pKa = 11.84TAGWYY285 pKa = 9.49RR286 pKa = 11.84LRR288 pKa = 11.84FQLVDD293 pKa = 3.4VQNRR297 pKa = 11.84GSPHH301 pKa = 6.52AFTRR305 pKa = 11.84SGLNLSKK312 pKa = 10.88CSLRR316 pKa = 11.84KK317 pKa = 9.66IFLGG321 pKa = 3.78
Molecular weight: 35.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4463529 |
30 |
5875 |
434.1 |
48.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.997 ± 0.019 | 1.229 ± 0.01 |
5.666 ± 0.016 | 6.296 ± 0.024 |
3.577 ± 0.015 | 6.594 ± 0.021 |
2.508 ± 0.009 | 5.043 ± 0.015 |
5.041 ± 0.022 | 8.813 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.078 ± 0.008 | 3.914 ± 0.013 |
6.311 ± 0.025 | 4.107 ± 0.019 |
6.576 ± 0.018 | 8.635 ± 0.027 |
5.901 ± 0.017 | 5.739 ± 0.015 |
1.304 ± 0.008 | 2.672 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
