
Tuhoko virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Rubulavirinae; Pararubulavirus; Tuhoko pararubulavirus 1
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
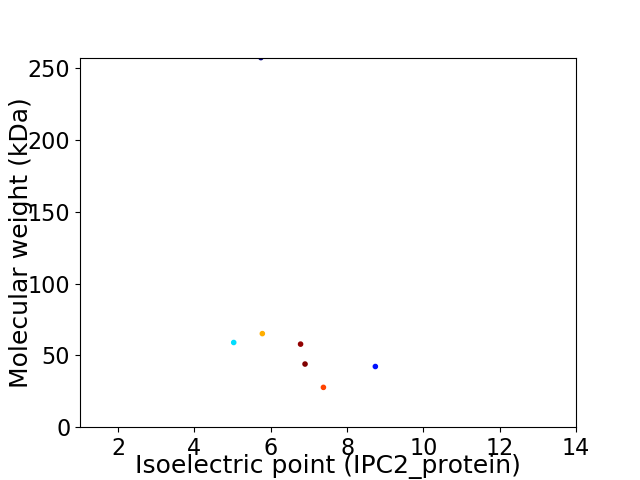
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D8WJ23|D8WJ23_9MONO Phosphoprotein OS=Tuhoko virus 1 OX=798072 GN=V/P PE=3 SV=1
MM1 pKa = 7.37SSVFKK6 pKa = 10.59AYY8 pKa = 10.12EE9 pKa = 3.97LFNLEE14 pKa = 4.0QEE16 pKa = 4.28QQEE19 pKa = 4.35RR20 pKa = 11.84GNDD23 pKa = 3.31LVLPPEE29 pKa = 4.21TLRR32 pKa = 11.84SNIRR36 pKa = 11.84VFVLNTQDD44 pKa = 3.22PEE46 pKa = 3.68IRR48 pKa = 11.84FRR50 pKa = 11.84MMCFCIRR57 pKa = 11.84LVASNSARR65 pKa = 11.84SAHH68 pKa = 5.69RR69 pKa = 11.84TGALLTLLSLPTAAMQNHH87 pKa = 5.79IRR89 pKa = 11.84IADD92 pKa = 4.23RR93 pKa = 11.84SPDD96 pKa = 3.44AEE98 pKa = 3.89IEE100 pKa = 4.18RR101 pKa = 11.84IEE103 pKa = 4.55IEE105 pKa = 4.11GFEE108 pKa = 3.96QGSYY112 pKa = 11.2KK113 pKa = 10.72LIPNARR119 pKa = 11.84TPMSPAEE126 pKa = 4.05ITALDD131 pKa = 4.05AMATDD136 pKa = 4.74LPDD139 pKa = 5.41SIMYY143 pKa = 7.57DD144 pKa = 3.54TPFMNMYY151 pKa = 9.44TEE153 pKa = 5.72AEE155 pKa = 4.22GCDD158 pKa = 3.7EE159 pKa = 4.16IEE161 pKa = 4.31EE162 pKa = 4.1FLEE165 pKa = 4.12AVYY168 pKa = 10.84SVLIQVWITVCKK180 pKa = 10.58CMTAYY185 pKa = 9.75DD186 pKa = 3.94QPTGSDD192 pKa = 3.55EE193 pKa = 4.19KK194 pKa = 11.32RR195 pKa = 11.84MAKK198 pKa = 9.77YY199 pKa = 9.33QQQGRR204 pKa = 11.84LDD206 pKa = 3.58PKK208 pKa = 9.71YY209 pKa = 9.02TLQQEE214 pKa = 4.37VRR216 pKa = 11.84RR217 pKa = 11.84LIQKK221 pKa = 8.65CIRR224 pKa = 11.84GSLPVRR230 pKa = 11.84QFLAFEE236 pKa = 4.33LQTARR241 pKa = 11.84KK242 pKa = 9.05QGTITSKK249 pKa = 9.51YY250 pKa = 8.2YY251 pKa = 11.52SMVGDD256 pKa = 3.56ISKK259 pKa = 10.72YY260 pKa = 9.72IEE262 pKa = 4.15NAGMSAFFMTARR274 pKa = 11.84YY275 pKa = 9.81ALGTRR280 pKa = 11.84WAPLALAAFTGDD292 pKa = 3.72LLKK295 pKa = 10.49MKK297 pKa = 10.44SLMLLYY303 pKa = 10.46RR304 pKa = 11.84KK305 pKa = 9.91LGEE308 pKa = 4.08KK309 pKa = 10.29ARR311 pKa = 11.84FMALLEE317 pKa = 4.48MPDD320 pKa = 3.33MMEE323 pKa = 4.7FAPANYY329 pKa = 9.99NLLYY333 pKa = 10.18SYY335 pKa = 11.64AMGIGSVMDD344 pKa = 3.55AQMRR348 pKa = 11.84NYY350 pKa = 10.61NFARR354 pKa = 11.84PFLNTAYY361 pKa = 10.01FQLGVEE367 pKa = 4.51TANKK371 pKa = 8.68QQGAVDD377 pKa = 3.57KK378 pKa = 11.61NMAAEE383 pKa = 4.73LGLSEE388 pKa = 4.75DD389 pKa = 4.66DD390 pKa = 3.88KK391 pKa = 11.62RR392 pKa = 11.84VMAQTVSRR400 pKa = 11.84LTGNKK405 pKa = 9.73AGSDD409 pKa = 3.77NQDD412 pKa = 3.25LMDD415 pKa = 3.83MLSRR419 pKa = 11.84RR420 pKa = 11.84SRR422 pKa = 11.84SGQTRR427 pKa = 11.84ALRR430 pKa = 11.84MSAQGNQAVDD440 pKa = 3.33GDD442 pKa = 4.18EE443 pKa = 4.41EE444 pKa = 5.09DD445 pKa = 4.91DD446 pKa = 3.89EE447 pKa = 4.83EE448 pKa = 4.73EE449 pKa = 4.31EE450 pKa = 4.32EE451 pKa = 4.19EE452 pKa = 4.49EE453 pKa = 4.15EE454 pKa = 5.12ARR456 pKa = 11.84DD457 pKa = 3.69LTEE460 pKa = 4.34EE461 pKa = 4.02EE462 pKa = 4.39QKK464 pKa = 11.04EE465 pKa = 4.21SEE467 pKa = 3.89AWLKK471 pKa = 9.53KK472 pKa = 8.6WKK474 pKa = 10.14EE475 pKa = 3.78IEE477 pKa = 4.23AQRR480 pKa = 11.84HH481 pKa = 3.87AAIEE485 pKa = 3.84AMKK488 pKa = 9.88KK489 pKa = 9.75HH490 pKa = 5.71KK491 pKa = 10.1AARR494 pKa = 11.84TQPSQMPSEE503 pKa = 4.24VQVHH507 pKa = 5.82NEE509 pKa = 3.94NQDD512 pKa = 3.05VRR514 pKa = 11.84GDD516 pKa = 3.61LDD518 pKa = 3.5SS519 pKa = 4.11
MM1 pKa = 7.37SSVFKK6 pKa = 10.59AYY8 pKa = 10.12EE9 pKa = 3.97LFNLEE14 pKa = 4.0QEE16 pKa = 4.28QQEE19 pKa = 4.35RR20 pKa = 11.84GNDD23 pKa = 3.31LVLPPEE29 pKa = 4.21TLRR32 pKa = 11.84SNIRR36 pKa = 11.84VFVLNTQDD44 pKa = 3.22PEE46 pKa = 3.68IRR48 pKa = 11.84FRR50 pKa = 11.84MMCFCIRR57 pKa = 11.84LVASNSARR65 pKa = 11.84SAHH68 pKa = 5.69RR69 pKa = 11.84TGALLTLLSLPTAAMQNHH87 pKa = 5.79IRR89 pKa = 11.84IADD92 pKa = 4.23RR93 pKa = 11.84SPDD96 pKa = 3.44AEE98 pKa = 3.89IEE100 pKa = 4.18RR101 pKa = 11.84IEE103 pKa = 4.55IEE105 pKa = 4.11GFEE108 pKa = 3.96QGSYY112 pKa = 11.2KK113 pKa = 10.72LIPNARR119 pKa = 11.84TPMSPAEE126 pKa = 4.05ITALDD131 pKa = 4.05AMATDD136 pKa = 4.74LPDD139 pKa = 5.41SIMYY143 pKa = 7.57DD144 pKa = 3.54TPFMNMYY151 pKa = 9.44TEE153 pKa = 5.72AEE155 pKa = 4.22GCDD158 pKa = 3.7EE159 pKa = 4.16IEE161 pKa = 4.31EE162 pKa = 4.1FLEE165 pKa = 4.12AVYY168 pKa = 10.84SVLIQVWITVCKK180 pKa = 10.58CMTAYY185 pKa = 9.75DD186 pKa = 3.94QPTGSDD192 pKa = 3.55EE193 pKa = 4.19KK194 pKa = 11.32RR195 pKa = 11.84MAKK198 pKa = 9.77YY199 pKa = 9.33QQQGRR204 pKa = 11.84LDD206 pKa = 3.58PKK208 pKa = 9.71YY209 pKa = 9.02TLQQEE214 pKa = 4.37VRR216 pKa = 11.84RR217 pKa = 11.84LIQKK221 pKa = 8.65CIRR224 pKa = 11.84GSLPVRR230 pKa = 11.84QFLAFEE236 pKa = 4.33LQTARR241 pKa = 11.84KK242 pKa = 9.05QGTITSKK249 pKa = 9.51YY250 pKa = 8.2YY251 pKa = 11.52SMVGDD256 pKa = 3.56ISKK259 pKa = 10.72YY260 pKa = 9.72IEE262 pKa = 4.15NAGMSAFFMTARR274 pKa = 11.84YY275 pKa = 9.81ALGTRR280 pKa = 11.84WAPLALAAFTGDD292 pKa = 3.72LLKK295 pKa = 10.49MKK297 pKa = 10.44SLMLLYY303 pKa = 10.46RR304 pKa = 11.84KK305 pKa = 9.91LGEE308 pKa = 4.08KK309 pKa = 10.29ARR311 pKa = 11.84FMALLEE317 pKa = 4.48MPDD320 pKa = 3.33MMEE323 pKa = 4.7FAPANYY329 pKa = 9.99NLLYY333 pKa = 10.18SYY335 pKa = 11.64AMGIGSVMDD344 pKa = 3.55AQMRR348 pKa = 11.84NYY350 pKa = 10.61NFARR354 pKa = 11.84PFLNTAYY361 pKa = 10.01FQLGVEE367 pKa = 4.51TANKK371 pKa = 8.68QQGAVDD377 pKa = 3.57KK378 pKa = 11.61NMAAEE383 pKa = 4.73LGLSEE388 pKa = 4.75DD389 pKa = 4.66DD390 pKa = 3.88KK391 pKa = 11.62RR392 pKa = 11.84VMAQTVSRR400 pKa = 11.84LTGNKK405 pKa = 9.73AGSDD409 pKa = 3.77NQDD412 pKa = 3.25LMDD415 pKa = 3.83MLSRR419 pKa = 11.84RR420 pKa = 11.84SRR422 pKa = 11.84SGQTRR427 pKa = 11.84ALRR430 pKa = 11.84MSAQGNQAVDD440 pKa = 3.33GDD442 pKa = 4.18EE443 pKa = 4.41EE444 pKa = 5.09DD445 pKa = 4.91DD446 pKa = 3.89EE447 pKa = 4.83EE448 pKa = 4.73EE449 pKa = 4.31EE450 pKa = 4.32EE451 pKa = 4.19EE452 pKa = 4.49EE453 pKa = 4.15EE454 pKa = 5.12ARR456 pKa = 11.84DD457 pKa = 3.69LTEE460 pKa = 4.34EE461 pKa = 4.02EE462 pKa = 4.39QKK464 pKa = 11.04EE465 pKa = 4.21SEE467 pKa = 3.89AWLKK471 pKa = 9.53KK472 pKa = 8.6WKK474 pKa = 10.14EE475 pKa = 3.78IEE477 pKa = 4.23AQRR480 pKa = 11.84HH481 pKa = 3.87AAIEE485 pKa = 3.84AMKK488 pKa = 9.88KK489 pKa = 9.75HH490 pKa = 5.71KK491 pKa = 10.1AARR494 pKa = 11.84TQPSQMPSEE503 pKa = 4.24VQVHH507 pKa = 5.82NEE509 pKa = 3.94NQDD512 pKa = 3.05VRR514 pKa = 11.84GDD516 pKa = 3.61LDD518 pKa = 3.5SS519 pKa = 4.11
Molecular weight: 59.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8WJ26|D8WJ26_9MONO Fusion glycoprotein F0 OS=Tuhoko virus 1 OX=798072 GN=F PE=3 SV=1
MM1 pKa = 7.74ASLIGASIPIPVNPDD16 pKa = 2.94SGKK19 pKa = 11.18SNLKK23 pKa = 9.36PFPIIKK29 pKa = 10.37APADD33 pKa = 3.65QSGHH37 pKa = 4.52TGKK40 pKa = 9.91IVRR43 pKa = 11.84QIRR46 pKa = 11.84IKK48 pKa = 10.91NLTPKK53 pKa = 10.38GSTEE57 pKa = 3.81IPITFMNTYY66 pKa = 10.74GFIKK70 pKa = 9.72PIWTHH75 pKa = 6.11GEE77 pKa = 3.97FFSEE81 pKa = 4.15FHH83 pKa = 6.95RR84 pKa = 11.84NNSTPSLTACMLPFGTGPFVDD105 pKa = 4.48NPSKK109 pKa = 11.07LLGDD113 pKa = 3.97LEE115 pKa = 4.71KK116 pKa = 10.74IQINVKK122 pKa = 10.0KK123 pKa = 10.24SASLRR128 pKa = 11.84EE129 pKa = 3.91EE130 pKa = 4.07VIFDD134 pKa = 3.38IKK136 pKa = 10.06TIPFAFTKK144 pKa = 9.86FQIGRR149 pKa = 11.84EE150 pKa = 3.91RR151 pKa = 11.84IICVSSDD158 pKa = 3.39RR159 pKa = 11.84YY160 pKa = 9.84VKK162 pKa = 10.85CPGKK166 pKa = 8.98LTSGVEE172 pKa = 3.81YY173 pKa = 9.79SYY175 pKa = 11.22CIAFISLTFCPEE187 pKa = 3.77SYY189 pKa = 10.27KK190 pKa = 10.63FRR192 pKa = 11.84VARR195 pKa = 11.84PLQQLRR201 pKa = 11.84TSYY204 pKa = 9.68MRR206 pKa = 11.84AIQLEE211 pKa = 4.56IIMKK215 pKa = 9.66IDD217 pKa = 3.93CAPDD221 pKa = 3.51SPIKK225 pKa = 10.65RR226 pKa = 11.84NLIYY230 pKa = 10.79DD231 pKa = 4.02KK232 pKa = 11.41EE233 pKa = 4.08NDD235 pKa = 3.17IYY237 pKa = 10.71IASVWLHH244 pKa = 6.24LCNILKK250 pKa = 10.35GKK252 pKa = 8.84NHH254 pKa = 6.73FKK256 pKa = 10.46TYY258 pKa = 10.64DD259 pKa = 3.2DD260 pKa = 3.96TYY262 pKa = 11.17FANKK266 pKa = 8.08CRR268 pKa = 11.84KK269 pKa = 7.54MQLVCGIADD278 pKa = 3.27MWGPTIIVHH287 pKa = 5.66SKK289 pKa = 10.27GRR291 pKa = 11.84IPKK294 pKa = 9.75SAAPYY299 pKa = 9.8FNSKK303 pKa = 8.39GWACHH308 pKa = 5.82PLSEE312 pKa = 4.46IAPSITKK319 pKa = 9.99ILWSVGATILQVNAVLQPSDD339 pKa = 3.18ISSAAGTSDD348 pKa = 5.18LIYY351 pKa = 10.09PKK353 pKa = 10.91VKK355 pKa = 10.31INPDD359 pKa = 3.2LAEE362 pKa = 3.92EE363 pKa = 5.32RR364 pKa = 11.84GIKK367 pKa = 8.86WNPLRR372 pKa = 11.84KK373 pKa = 9.16AVHH376 pKa = 5.25STT378 pKa = 3.02
MM1 pKa = 7.74ASLIGASIPIPVNPDD16 pKa = 2.94SGKK19 pKa = 11.18SNLKK23 pKa = 9.36PFPIIKK29 pKa = 10.37APADD33 pKa = 3.65QSGHH37 pKa = 4.52TGKK40 pKa = 9.91IVRR43 pKa = 11.84QIRR46 pKa = 11.84IKK48 pKa = 10.91NLTPKK53 pKa = 10.38GSTEE57 pKa = 3.81IPITFMNTYY66 pKa = 10.74GFIKK70 pKa = 9.72PIWTHH75 pKa = 6.11GEE77 pKa = 3.97FFSEE81 pKa = 4.15FHH83 pKa = 6.95RR84 pKa = 11.84NNSTPSLTACMLPFGTGPFVDD105 pKa = 4.48NPSKK109 pKa = 11.07LLGDD113 pKa = 3.97LEE115 pKa = 4.71KK116 pKa = 10.74IQINVKK122 pKa = 10.0KK123 pKa = 10.24SASLRR128 pKa = 11.84EE129 pKa = 3.91EE130 pKa = 4.07VIFDD134 pKa = 3.38IKK136 pKa = 10.06TIPFAFTKK144 pKa = 9.86FQIGRR149 pKa = 11.84EE150 pKa = 3.91RR151 pKa = 11.84IICVSSDD158 pKa = 3.39RR159 pKa = 11.84YY160 pKa = 9.84VKK162 pKa = 10.85CPGKK166 pKa = 8.98LTSGVEE172 pKa = 3.81YY173 pKa = 9.79SYY175 pKa = 11.22CIAFISLTFCPEE187 pKa = 3.77SYY189 pKa = 10.27KK190 pKa = 10.63FRR192 pKa = 11.84VARR195 pKa = 11.84PLQQLRR201 pKa = 11.84TSYY204 pKa = 9.68MRR206 pKa = 11.84AIQLEE211 pKa = 4.56IIMKK215 pKa = 9.66IDD217 pKa = 3.93CAPDD221 pKa = 3.51SPIKK225 pKa = 10.65RR226 pKa = 11.84NLIYY230 pKa = 10.79DD231 pKa = 4.02KK232 pKa = 11.41EE233 pKa = 4.08NDD235 pKa = 3.17IYY237 pKa = 10.71IASVWLHH244 pKa = 6.24LCNILKK250 pKa = 10.35GKK252 pKa = 8.84NHH254 pKa = 6.73FKK256 pKa = 10.46TYY258 pKa = 10.64DD259 pKa = 3.2DD260 pKa = 3.96TYY262 pKa = 11.17FANKK266 pKa = 8.08CRR268 pKa = 11.84KK269 pKa = 7.54MQLVCGIADD278 pKa = 3.27MWGPTIIVHH287 pKa = 5.66SKK289 pKa = 10.27GRR291 pKa = 11.84IPKK294 pKa = 9.75SAAPYY299 pKa = 9.8FNSKK303 pKa = 8.39GWACHH308 pKa = 5.82PLSEE312 pKa = 4.46IAPSITKK319 pKa = 9.99ILWSVGATILQVNAVLQPSDD339 pKa = 3.18ISSAAGTSDD348 pKa = 5.18LIYY351 pKa = 10.09PKK353 pKa = 10.91VKK355 pKa = 10.31INPDD359 pKa = 3.2LAEE362 pKa = 3.92EE363 pKa = 5.32RR364 pKa = 11.84GIKK367 pKa = 8.86WNPLRR372 pKa = 11.84KK373 pKa = 9.16AVHH376 pKa = 5.25STT378 pKa = 3.02
Molecular weight: 42.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4944 |
252 |
2274 |
706.3 |
79.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.655 ± 0.885 | 1.922 ± 0.289 |
5.239 ± 0.3 | 5.057 ± 0.637 |
3.236 ± 0.26 | 4.511 ± 0.306 |
2.124 ± 0.333 | 8.212 ± 0.737 |
5.218 ± 0.523 | 10.558 ± 1.375 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.447 ± 0.502 | 5.178 ± 0.416 |
5.502 ± 0.829 | 4.956 ± 0.447 |
4.49 ± 0.386 | 8.657 ± 0.403 |
6.29 ± 0.384 | 5.259 ± 0.315 |
1.032 ± 0.185 | 3.459 ± 0.483 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
