
Bacteriophage sp.
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
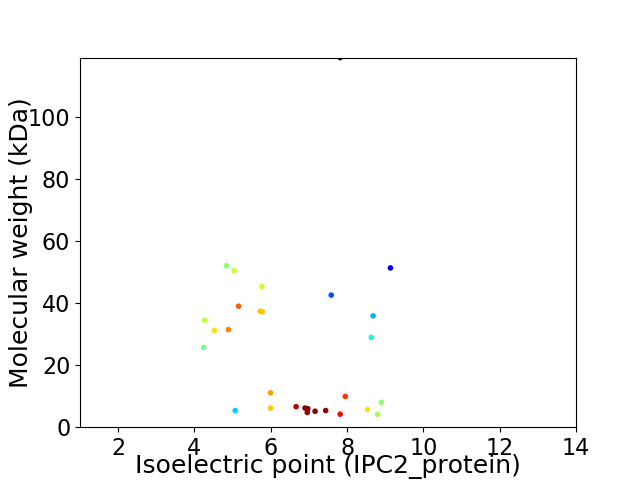
Virtual 2D-PAGE plot for 29 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
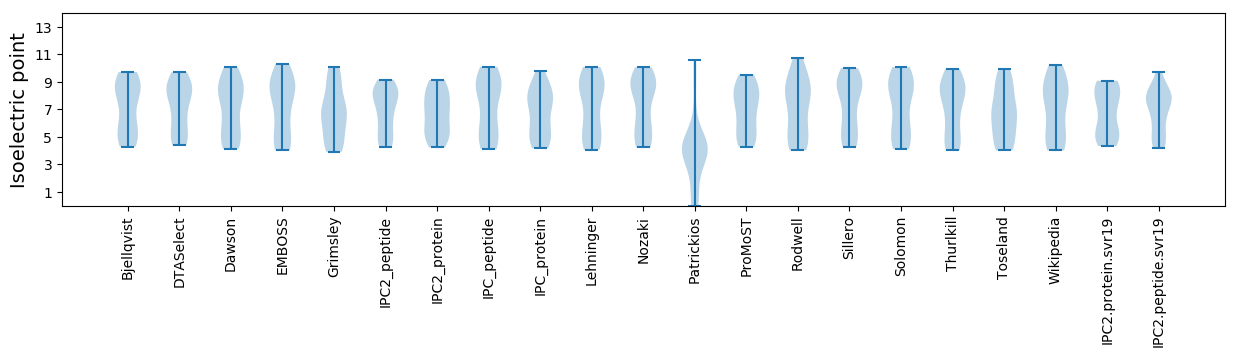
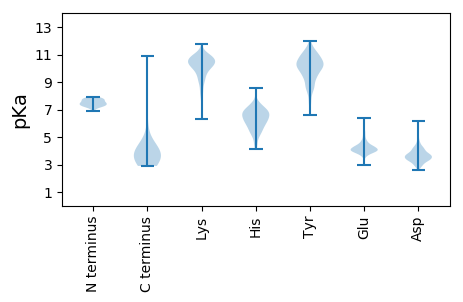
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H7BVF6|H7BVF6_9VIRU Uncharacterized protein OS=Bacteriophage sp. OX=38018 GN=2019_scaffold132_00094 PE=4 SV=1
MM1 pKa = 7.55WYY3 pKa = 10.37VYY5 pKa = 10.21TDD7 pKa = 3.62PTSGSQYY14 pKa = 10.66VFDD17 pKa = 4.33TSEE20 pKa = 3.8VMHH23 pKa = 6.77FKK25 pKa = 10.46TSFSFDD31 pKa = 3.3GVTGLPVQQILRR43 pKa = 11.84DD44 pKa = 4.12TISGASASQRR54 pKa = 11.84YY55 pKa = 7.49MNSLYY60 pKa = 10.76EE61 pKa = 4.3SGLTAKK67 pKa = 9.39ATLEE71 pKa = 4.04YY72 pKa = 10.37TGEE75 pKa = 4.19LNDD78 pKa = 3.8KK79 pKa = 10.81AKK81 pKa = 10.39EE82 pKa = 3.82ALVKK86 pKa = 10.72SFEE89 pKa = 4.53DD90 pKa = 3.81FGSGARR96 pKa = 11.84NTGKK100 pKa = 10.43IIPVPLGMKK109 pKa = 7.67LTPLDD114 pKa = 3.64IKK116 pKa = 11.16LSDD119 pKa = 3.56SQFFEE124 pKa = 4.3LKK126 pKa = 10.31KK127 pKa = 10.06YY128 pKa = 8.25TALQIAAAFGVKK140 pKa = 9.71PNQINDD146 pKa = 3.6YY147 pKa = 10.4SKK149 pKa = 10.95SSYY152 pKa = 11.28ANSEE156 pKa = 3.97LQQLSFYY163 pKa = 11.13VDD165 pKa = 3.13TEE167 pKa = 4.29LFVIKK172 pKa = 10.31QYY174 pKa = 10.56EE175 pKa = 3.9EE176 pKa = 4.05EE177 pKa = 4.25INYY180 pKa = 10.24KK181 pKa = 9.41MLTDD185 pKa = 4.18EE186 pKa = 4.42EE187 pKa = 4.43QDD189 pKa = 3.1DD190 pKa = 4.29GFYY193 pKa = 11.06YY194 pKa = 10.52KK195 pKa = 10.95YY196 pKa = 10.49NEE198 pKa = 4.11KK199 pKa = 10.81VLFRR203 pKa = 11.84TDD205 pKa = 3.02SKK207 pKa = 10.19TQMEE211 pKa = 4.39YY212 pKa = 11.08LKK214 pKa = 10.72NGVSGSIMKK223 pKa = 10.07PNEE226 pKa = 3.52ARR228 pKa = 11.84RR229 pKa = 11.84KK230 pKa = 9.88LDD232 pKa = 3.66LPDD235 pKa = 4.81GEE237 pKa = 5.86GGDD240 pKa = 3.77TLLANGSIVPLTMAGAAYY258 pKa = 10.07QKK260 pKa = 10.95GQIEE264 pKa = 4.2QEE266 pKa = 4.16EE267 pKa = 4.87TEE269 pKa = 4.48KK270 pKa = 11.03PEE272 pKa = 3.94QPEE275 pKa = 4.3EE276 pKa = 3.93EE277 pKa = 4.49TEE279 pKa = 4.22PDD281 pKa = 3.4TEE283 pKa = 4.41QPDD286 pKa = 3.89TTGQPDD292 pKa = 3.62EE293 pKa = 4.24TDD295 pKa = 3.13EE296 pKa = 5.91AEE298 pKa = 4.95DD299 pKa = 3.96EE300 pKa = 4.32EE301 pKa = 4.41EE302 pKa = 4.55QEE304 pKa = 4.4GGEE307 pKa = 4.12
MM1 pKa = 7.55WYY3 pKa = 10.37VYY5 pKa = 10.21TDD7 pKa = 3.62PTSGSQYY14 pKa = 10.66VFDD17 pKa = 4.33TSEE20 pKa = 3.8VMHH23 pKa = 6.77FKK25 pKa = 10.46TSFSFDD31 pKa = 3.3GVTGLPVQQILRR43 pKa = 11.84DD44 pKa = 4.12TISGASASQRR54 pKa = 11.84YY55 pKa = 7.49MNSLYY60 pKa = 10.76EE61 pKa = 4.3SGLTAKK67 pKa = 9.39ATLEE71 pKa = 4.04YY72 pKa = 10.37TGEE75 pKa = 4.19LNDD78 pKa = 3.8KK79 pKa = 10.81AKK81 pKa = 10.39EE82 pKa = 3.82ALVKK86 pKa = 10.72SFEE89 pKa = 4.53DD90 pKa = 3.81FGSGARR96 pKa = 11.84NTGKK100 pKa = 10.43IIPVPLGMKK109 pKa = 7.67LTPLDD114 pKa = 3.64IKK116 pKa = 11.16LSDD119 pKa = 3.56SQFFEE124 pKa = 4.3LKK126 pKa = 10.31KK127 pKa = 10.06YY128 pKa = 8.25TALQIAAAFGVKK140 pKa = 9.71PNQINDD146 pKa = 3.6YY147 pKa = 10.4SKK149 pKa = 10.95SSYY152 pKa = 11.28ANSEE156 pKa = 3.97LQQLSFYY163 pKa = 11.13VDD165 pKa = 3.13TEE167 pKa = 4.29LFVIKK172 pKa = 10.31QYY174 pKa = 10.56EE175 pKa = 3.9EE176 pKa = 4.05EE177 pKa = 4.25INYY180 pKa = 10.24KK181 pKa = 9.41MLTDD185 pKa = 4.18EE186 pKa = 4.42EE187 pKa = 4.43QDD189 pKa = 3.1DD190 pKa = 4.29GFYY193 pKa = 11.06YY194 pKa = 10.52KK195 pKa = 10.95YY196 pKa = 10.49NEE198 pKa = 4.11KK199 pKa = 10.81VLFRR203 pKa = 11.84TDD205 pKa = 3.02SKK207 pKa = 10.19TQMEE211 pKa = 4.39YY212 pKa = 11.08LKK214 pKa = 10.72NGVSGSIMKK223 pKa = 10.07PNEE226 pKa = 3.52ARR228 pKa = 11.84RR229 pKa = 11.84KK230 pKa = 9.88LDD232 pKa = 3.66LPDD235 pKa = 4.81GEE237 pKa = 5.86GGDD240 pKa = 3.77TLLANGSIVPLTMAGAAYY258 pKa = 10.07QKK260 pKa = 10.95GQIEE264 pKa = 4.2QEE266 pKa = 4.16EE267 pKa = 4.87TEE269 pKa = 4.48KK270 pKa = 11.03PEE272 pKa = 3.94QPEE275 pKa = 4.3EE276 pKa = 3.93EE277 pKa = 4.49TEE279 pKa = 4.22PDD281 pKa = 3.4TEE283 pKa = 4.41QPDD286 pKa = 3.89TTGQPDD292 pKa = 3.62EE293 pKa = 4.24TDD295 pKa = 3.13EE296 pKa = 5.91AEE298 pKa = 4.95DD299 pKa = 3.96EE300 pKa = 4.32EE301 pKa = 4.41EE302 pKa = 4.55QEE304 pKa = 4.4GGEE307 pKa = 4.12
Molecular weight: 34.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H7BVF0|H7BVF0_9VIRU Uncharacterized protein OS=Bacteriophage sp. OX=38018 GN=2019_scaffold132_00065 PE=4 SV=1
MM1 pKa = 7.42GKK3 pKa = 10.14KK4 pKa = 9.73SVNNLYY10 pKa = 10.79KK11 pKa = 10.75PMLEE15 pKa = 4.19HH16 pKa = 7.33SNVEE20 pKa = 4.28QKK22 pKa = 9.6FHH24 pKa = 6.86KK25 pKa = 9.78AAKK28 pKa = 9.9GKK30 pKa = 7.47TEE32 pKa = 3.98RR33 pKa = 11.84PDD35 pKa = 3.1VAVILEE41 pKa = 3.98PTNIQRR47 pKa = 11.84HH48 pKa = 4.44VKK50 pKa = 10.21NVIEE54 pKa = 4.05QLEE57 pKa = 4.23NTAPEE62 pKa = 5.42GYY64 pKa = 10.01DD65 pKa = 3.26VPHH68 pKa = 7.3PEE70 pKa = 4.37KK71 pKa = 9.9AWKK74 pKa = 9.12PSRR77 pKa = 11.84HH78 pKa = 5.32GKK80 pKa = 8.94VCINEE85 pKa = 3.98GTSRR89 pKa = 11.84KK90 pKa = 8.89VRR92 pKa = 11.84MIEE95 pKa = 3.81KK96 pKa = 9.67PRR98 pKa = 11.84YY99 pKa = 8.55NYY101 pKa = 9.96EE102 pKa = 3.9QVIHH106 pKa = 6.74HH107 pKa = 6.44IVVSACYY114 pKa = 9.98DD115 pKa = 2.95IFMKK119 pKa = 10.55GMYY122 pKa = 9.2EE123 pKa = 4.13FSCGSVPNRR132 pKa = 11.84GAHH135 pKa = 5.31YY136 pKa = 8.87GKK138 pKa = 10.44KK139 pKa = 10.42YY140 pKa = 8.75IEE142 pKa = 3.89RR143 pKa = 11.84WIQRR147 pKa = 11.84DD148 pKa = 3.55KK149 pKa = 11.51KK150 pKa = 8.71NCKK153 pKa = 9.57YY154 pKa = 9.85VLKK157 pKa = 10.08MDD159 pKa = 2.97IRR161 pKa = 11.84HH162 pKa = 5.61FFEE165 pKa = 5.89SVDD168 pKa = 3.64HH169 pKa = 7.2DD170 pKa = 4.43VLKK173 pKa = 11.0AWLKK177 pKa = 10.74KK178 pKa = 10.13KK179 pKa = 10.38IRR181 pKa = 11.84DD182 pKa = 3.38EE183 pKa = 4.2RR184 pKa = 11.84MLYY187 pKa = 9.97ILEE190 pKa = 4.83LIIDD194 pKa = 4.34GSEE197 pKa = 3.68VGLPLGFYY205 pKa = 9.61TSQWLSNFMLQPLDD219 pKa = 3.98HH220 pKa = 7.2FIKK223 pKa = 10.05EE224 pKa = 3.93QLKK227 pKa = 8.12AVHH230 pKa = 6.71YY231 pKa = 9.66IRR233 pKa = 11.84YY234 pKa = 7.55MDD236 pKa = 4.14DD237 pKa = 2.87MVVFGKK243 pKa = 10.11NKK245 pKa = 10.13KK246 pKa = 8.46EE247 pKa = 3.84LHH249 pKa = 6.18RR250 pKa = 11.84MQQEE254 pKa = 3.55IEE256 pKa = 3.91RR257 pKa = 11.84YY258 pKa = 9.42LRR260 pKa = 11.84EE261 pKa = 4.04KK262 pKa = 10.94FNLQMKK268 pKa = 9.27GNWQVFRR275 pKa = 11.84FDD277 pKa = 3.48YY278 pKa = 10.52TEE280 pKa = 3.94KK281 pKa = 10.03KK282 pKa = 7.4TGKK285 pKa = 10.06RR286 pKa = 11.84KK287 pKa = 9.52GRR289 pKa = 11.84PLDD292 pKa = 3.48FMGFQFYY299 pKa = 10.39HH300 pKa = 7.45DD301 pKa = 3.7KK302 pKa = 10.81TILRR306 pKa = 11.84EE307 pKa = 4.29SIMLSCTRR315 pKa = 11.84KK316 pKa = 9.28VNRR319 pKa = 11.84VAKK322 pKa = 10.0KK323 pKa = 10.54EE324 pKa = 3.97KK325 pKa = 8.97ITWYY329 pKa = 10.03DD330 pKa = 2.9ATAILSYY337 pKa = 9.84MGYY340 pKa = 10.76LSNTDD345 pKa = 3.25TYY347 pKa = 12.01DD348 pKa = 3.0MYY350 pKa = 11.21LQRR353 pKa = 11.84VKK355 pKa = 10.63PYY357 pKa = 11.34VNVKK361 pKa = 9.43KK362 pKa = 10.61LKK364 pKa = 10.48KK365 pKa = 9.67IVSKK369 pKa = 10.52HH370 pKa = 4.39SKK372 pKa = 8.58RR373 pKa = 11.84KK374 pKa = 8.75EE375 pKa = 3.98RR376 pKa = 11.84EE377 pKa = 3.37KK378 pKa = 10.62HH379 pKa = 4.49EE380 pKa = 4.08RR381 pKa = 11.84MEE383 pKa = 4.12RR384 pKa = 11.84SVRR387 pKa = 11.84NGGRR391 pKa = 11.84TAGGVRR397 pKa = 11.84HH398 pKa = 6.46NSITDD403 pKa = 3.36NGVSEE408 pKa = 4.32TQYY411 pKa = 10.76QEE413 pKa = 4.04SNEE416 pKa = 4.06RR417 pKa = 11.84GCRR420 pKa = 11.84RR421 pKa = 11.84EE422 pKa = 3.68EE423 pKa = 4.01SHH425 pKa = 7.1RR426 pKa = 11.84MAARR430 pKa = 11.84GAA432 pKa = 3.52
MM1 pKa = 7.42GKK3 pKa = 10.14KK4 pKa = 9.73SVNNLYY10 pKa = 10.79KK11 pKa = 10.75PMLEE15 pKa = 4.19HH16 pKa = 7.33SNVEE20 pKa = 4.28QKK22 pKa = 9.6FHH24 pKa = 6.86KK25 pKa = 9.78AAKK28 pKa = 9.9GKK30 pKa = 7.47TEE32 pKa = 3.98RR33 pKa = 11.84PDD35 pKa = 3.1VAVILEE41 pKa = 3.98PTNIQRR47 pKa = 11.84HH48 pKa = 4.44VKK50 pKa = 10.21NVIEE54 pKa = 4.05QLEE57 pKa = 4.23NTAPEE62 pKa = 5.42GYY64 pKa = 10.01DD65 pKa = 3.26VPHH68 pKa = 7.3PEE70 pKa = 4.37KK71 pKa = 9.9AWKK74 pKa = 9.12PSRR77 pKa = 11.84HH78 pKa = 5.32GKK80 pKa = 8.94VCINEE85 pKa = 3.98GTSRR89 pKa = 11.84KK90 pKa = 8.89VRR92 pKa = 11.84MIEE95 pKa = 3.81KK96 pKa = 9.67PRR98 pKa = 11.84YY99 pKa = 8.55NYY101 pKa = 9.96EE102 pKa = 3.9QVIHH106 pKa = 6.74HH107 pKa = 6.44IVVSACYY114 pKa = 9.98DD115 pKa = 2.95IFMKK119 pKa = 10.55GMYY122 pKa = 9.2EE123 pKa = 4.13FSCGSVPNRR132 pKa = 11.84GAHH135 pKa = 5.31YY136 pKa = 8.87GKK138 pKa = 10.44KK139 pKa = 10.42YY140 pKa = 8.75IEE142 pKa = 3.89RR143 pKa = 11.84WIQRR147 pKa = 11.84DD148 pKa = 3.55KK149 pKa = 11.51KK150 pKa = 8.71NCKK153 pKa = 9.57YY154 pKa = 9.85VLKK157 pKa = 10.08MDD159 pKa = 2.97IRR161 pKa = 11.84HH162 pKa = 5.61FFEE165 pKa = 5.89SVDD168 pKa = 3.64HH169 pKa = 7.2DD170 pKa = 4.43VLKK173 pKa = 11.0AWLKK177 pKa = 10.74KK178 pKa = 10.13KK179 pKa = 10.38IRR181 pKa = 11.84DD182 pKa = 3.38EE183 pKa = 4.2RR184 pKa = 11.84MLYY187 pKa = 9.97ILEE190 pKa = 4.83LIIDD194 pKa = 4.34GSEE197 pKa = 3.68VGLPLGFYY205 pKa = 9.61TSQWLSNFMLQPLDD219 pKa = 3.98HH220 pKa = 7.2FIKK223 pKa = 10.05EE224 pKa = 3.93QLKK227 pKa = 8.12AVHH230 pKa = 6.71YY231 pKa = 9.66IRR233 pKa = 11.84YY234 pKa = 7.55MDD236 pKa = 4.14DD237 pKa = 2.87MVVFGKK243 pKa = 10.11NKK245 pKa = 10.13KK246 pKa = 8.46EE247 pKa = 3.84LHH249 pKa = 6.18RR250 pKa = 11.84MQQEE254 pKa = 3.55IEE256 pKa = 3.91RR257 pKa = 11.84YY258 pKa = 9.42LRR260 pKa = 11.84EE261 pKa = 4.04KK262 pKa = 10.94FNLQMKK268 pKa = 9.27GNWQVFRR275 pKa = 11.84FDD277 pKa = 3.48YY278 pKa = 10.52TEE280 pKa = 3.94KK281 pKa = 10.03KK282 pKa = 7.4TGKK285 pKa = 10.06RR286 pKa = 11.84KK287 pKa = 9.52GRR289 pKa = 11.84PLDD292 pKa = 3.48FMGFQFYY299 pKa = 10.39HH300 pKa = 7.45DD301 pKa = 3.7KK302 pKa = 10.81TILRR306 pKa = 11.84EE307 pKa = 4.29SIMLSCTRR315 pKa = 11.84KK316 pKa = 9.28VNRR319 pKa = 11.84VAKK322 pKa = 10.0KK323 pKa = 10.54EE324 pKa = 3.97KK325 pKa = 8.97ITWYY329 pKa = 10.03DD330 pKa = 2.9ATAILSYY337 pKa = 9.84MGYY340 pKa = 10.76LSNTDD345 pKa = 3.25TYY347 pKa = 12.01DD348 pKa = 3.0MYY350 pKa = 11.21LQRR353 pKa = 11.84VKK355 pKa = 10.63PYY357 pKa = 11.34VNVKK361 pKa = 9.43KK362 pKa = 10.61LKK364 pKa = 10.48KK365 pKa = 9.67IVSKK369 pKa = 10.52HH370 pKa = 4.39SKK372 pKa = 8.58RR373 pKa = 11.84KK374 pKa = 8.75EE375 pKa = 3.98RR376 pKa = 11.84EE377 pKa = 3.37KK378 pKa = 10.62HH379 pKa = 4.49EE380 pKa = 4.08RR381 pKa = 11.84MEE383 pKa = 4.12RR384 pKa = 11.84SVRR387 pKa = 11.84NGGRR391 pKa = 11.84TAGGVRR397 pKa = 11.84HH398 pKa = 6.46NSITDD403 pKa = 3.36NGVSEE408 pKa = 4.32TQYY411 pKa = 10.76QEE413 pKa = 4.04SNEE416 pKa = 4.06RR417 pKa = 11.84GCRR420 pKa = 11.84RR421 pKa = 11.84EE422 pKa = 3.68EE423 pKa = 4.01SHH425 pKa = 7.1RR426 pKa = 11.84MAARR430 pKa = 11.84GAA432 pKa = 3.52
Molecular weight: 51.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6670 |
36 |
1117 |
230.0 |
25.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.391 ± 0.995 | 1.874 ± 0.404 |
5.847 ± 0.548 | 7.721 ± 0.571 |
4.078 ± 0.369 | 6.492 ± 0.438 |
1.694 ± 0.339 | 5.847 ± 0.355 |
8.291 ± 0.663 | 7.286 ± 0.372 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.999 ± 0.207 | 4.798 ± 0.271 |
3.178 ± 0.466 | 3.568 ± 0.248 |
4.303 ± 0.73 | 6.672 ± 0.599 |
6.867 ± 0.883 | 6.117 ± 0.341 |
1.139 ± 0.155 | 3.838 ± 0.511 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
