
Luteimonas aestuarii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Luteimonas
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
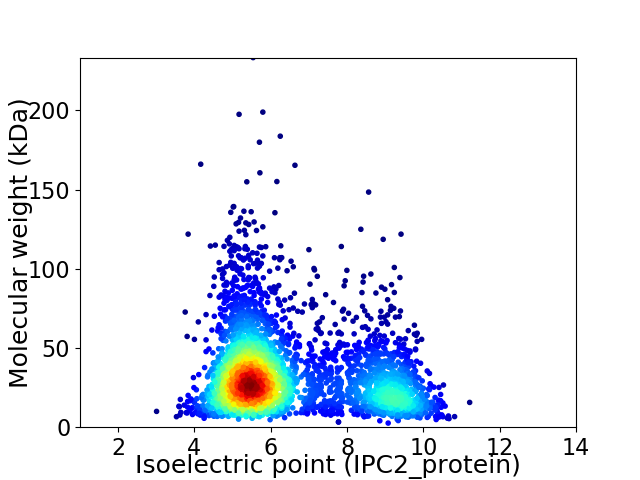
Virtual 2D-PAGE plot for 3335 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R5U4Q5|A0A4R5U4Q5_9GAMM Right-handed parallel beta-helix repeat-containing protein OS=Luteimonas aestuarii OX=453837 GN=E2F46_00415 PE=4 SV=1
MM1 pKa = 7.49TIKK4 pKa = 9.97TYY6 pKa = 10.4TPAEE10 pKa = 4.59AIQAAQAEE18 pKa = 4.77GCIEE22 pKa = 3.89IAAGVHH28 pKa = 6.52LSSQEE33 pKa = 4.08NIVADD38 pKa = 3.58QDD40 pKa = 3.6DD41 pKa = 4.03WSVEE45 pKa = 3.64GDD47 pKa = 3.44QMHH50 pKa = 7.74DD51 pKa = 3.63FTKK54 pKa = 10.47APYY57 pKa = 9.58WITTNDD63 pKa = 3.53GQVQPIYY70 pKa = 11.44GMDD73 pKa = 3.97DD74 pKa = 3.34KK75 pKa = 11.84DD76 pKa = 5.72LIDD79 pKa = 3.72VLANAA84 pKa = 5.26
MM1 pKa = 7.49TIKK4 pKa = 9.97TYY6 pKa = 10.4TPAEE10 pKa = 4.59AIQAAQAEE18 pKa = 4.77GCIEE22 pKa = 3.89IAAGVHH28 pKa = 6.52LSSQEE33 pKa = 4.08NIVADD38 pKa = 3.58QDD40 pKa = 3.6DD41 pKa = 4.03WSVEE45 pKa = 3.64GDD47 pKa = 3.44QMHH50 pKa = 7.74DD51 pKa = 3.63FTKK54 pKa = 10.47APYY57 pKa = 9.58WITTNDD63 pKa = 3.53GQVQPIYY70 pKa = 11.44GMDD73 pKa = 3.97DD74 pKa = 3.34KK75 pKa = 11.84DD76 pKa = 5.72LIDD79 pKa = 3.72VLANAA84 pKa = 5.26
Molecular weight: 9.19 kDa
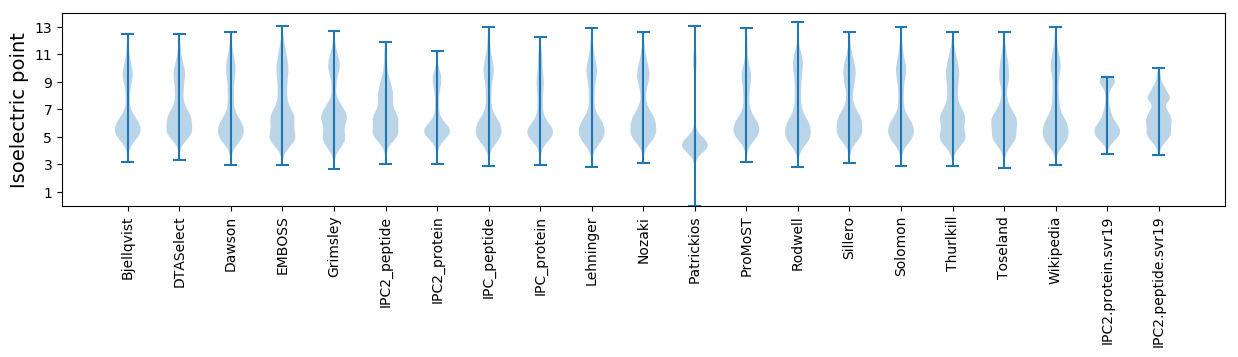
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R5TXZ5|A0A4R5TXZ5_9GAMM MPT synthase subunit 2 OS=Luteimonas aestuarii OX=453837 GN=E2F46_05525 PE=3 SV=1
MM1 pKa = 6.08TQARR5 pKa = 11.84RR6 pKa = 11.84TRR8 pKa = 11.84SKK10 pKa = 10.45EE11 pKa = 3.76RR12 pKa = 11.84GNGRR16 pKa = 11.84PAPPAAADD24 pKa = 3.42ARR26 pKa = 11.84EE27 pKa = 4.05AGLLYY32 pKa = 10.61VDD34 pKa = 5.49DD35 pKa = 4.69SAPGIHH41 pKa = 7.0RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84CGRR47 pKa = 11.84GFRR50 pKa = 11.84YY51 pKa = 10.39DD52 pKa = 3.87DD53 pKa = 3.41ATGVCIRR60 pKa = 11.84DD61 pKa = 3.38EE62 pKa = 4.16ATLARR67 pKa = 11.84IRR69 pKa = 11.84ALAIPPAYY77 pKa = 10.14VDD79 pKa = 3.15VWICASPRR87 pKa = 11.84GHH89 pKa = 6.67LQATGRR95 pKa = 11.84DD96 pKa = 3.29ARR98 pKa = 11.84GRR100 pKa = 11.84KK101 pKa = 7.65QYY103 pKa = 10.33RR104 pKa = 11.84YY105 pKa = 9.59HH106 pKa = 6.99PDD108 pKa = 2.62WRR110 pKa = 11.84STRR113 pKa = 11.84DD114 pKa = 2.81ADD116 pKa = 4.39KK117 pKa = 10.68FDD119 pKa = 4.01RR120 pKa = 11.84LLAFGKK126 pKa = 10.15ALPRR130 pKa = 11.84LRR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84LRR136 pKa = 11.84IDD138 pKa = 3.48LSLRR142 pKa = 11.84GLPRR146 pKa = 11.84EE147 pKa = 4.0KK148 pKa = 10.74ALAALVTIMATTLMRR163 pKa = 11.84IGNDD167 pKa = 2.6EE168 pKa = 3.99YY169 pKa = 11.64ARR171 pKa = 11.84DD172 pKa = 3.74NGSYY176 pKa = 11.32GLTTLRR182 pKa = 11.84NRR184 pKa = 11.84HH185 pKa = 5.46AKK187 pKa = 9.52FLRR190 pKa = 11.84DD191 pKa = 3.45GLHH194 pKa = 6.18LQFKK198 pKa = 9.93GKK200 pKa = 10.13GGKK203 pKa = 6.86THH205 pKa = 7.65DD206 pKa = 4.04AGIDD210 pKa = 3.3DD211 pKa = 4.07RR212 pKa = 11.84RR213 pKa = 11.84LVKK216 pKa = 10.46IMRR219 pKa = 11.84AMHH222 pKa = 6.49EE223 pKa = 4.27LPGQRR228 pKa = 11.84LFQYY232 pKa = 10.7RR233 pKa = 11.84DD234 pKa = 3.28DD235 pKa = 4.38EE236 pKa = 5.0GNLRR240 pKa = 11.84PLDD243 pKa = 3.87SSDD246 pKa = 3.4ANDD249 pKa = 3.66YY250 pKa = 10.83LRR252 pKa = 11.84EE253 pKa = 3.98RR254 pKa = 11.84MGGPFTAKK262 pKa = 10.49DD263 pKa = 3.53FRR265 pKa = 11.84TWGATATAFRR275 pKa = 11.84LLAATPVPEE284 pKa = 5.11DD285 pKa = 3.13ATKK288 pKa = 10.62RR289 pKa = 11.84DD290 pKa = 3.35IAGVEE295 pKa = 3.71RR296 pKa = 11.84DD297 pKa = 3.76VVNAVAEE304 pKa = 4.27MLGNTPAVCRR314 pKa = 11.84KK315 pKa = 10.15SYY317 pKa = 10.75IDD319 pKa = 3.38PCVFDD324 pKa = 3.77GWQDD328 pKa = 3.43GRR330 pKa = 11.84LHH332 pKa = 6.97RR333 pKa = 11.84AAQGARR339 pKa = 11.84GPRR342 pKa = 11.84QWEE345 pKa = 4.15TATLRR350 pKa = 11.84YY351 pKa = 9.05LRR353 pKa = 11.84RR354 pKa = 11.84VKK356 pKa = 10.62KK357 pKa = 10.17PSRR360 pKa = 3.51
MM1 pKa = 6.08TQARR5 pKa = 11.84RR6 pKa = 11.84TRR8 pKa = 11.84SKK10 pKa = 10.45EE11 pKa = 3.76RR12 pKa = 11.84GNGRR16 pKa = 11.84PAPPAAADD24 pKa = 3.42ARR26 pKa = 11.84EE27 pKa = 4.05AGLLYY32 pKa = 10.61VDD34 pKa = 5.49DD35 pKa = 4.69SAPGIHH41 pKa = 7.0RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84CGRR47 pKa = 11.84GFRR50 pKa = 11.84YY51 pKa = 10.39DD52 pKa = 3.87DD53 pKa = 3.41ATGVCIRR60 pKa = 11.84DD61 pKa = 3.38EE62 pKa = 4.16ATLARR67 pKa = 11.84IRR69 pKa = 11.84ALAIPPAYY77 pKa = 10.14VDD79 pKa = 3.15VWICASPRR87 pKa = 11.84GHH89 pKa = 6.67LQATGRR95 pKa = 11.84DD96 pKa = 3.29ARR98 pKa = 11.84GRR100 pKa = 11.84KK101 pKa = 7.65QYY103 pKa = 10.33RR104 pKa = 11.84YY105 pKa = 9.59HH106 pKa = 6.99PDD108 pKa = 2.62WRR110 pKa = 11.84STRR113 pKa = 11.84DD114 pKa = 2.81ADD116 pKa = 4.39KK117 pKa = 10.68FDD119 pKa = 4.01RR120 pKa = 11.84LLAFGKK126 pKa = 10.15ALPRR130 pKa = 11.84LRR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84LRR136 pKa = 11.84IDD138 pKa = 3.48LSLRR142 pKa = 11.84GLPRR146 pKa = 11.84EE147 pKa = 4.0KK148 pKa = 10.74ALAALVTIMATTLMRR163 pKa = 11.84IGNDD167 pKa = 2.6EE168 pKa = 3.99YY169 pKa = 11.64ARR171 pKa = 11.84DD172 pKa = 3.74NGSYY176 pKa = 11.32GLTTLRR182 pKa = 11.84NRR184 pKa = 11.84HH185 pKa = 5.46AKK187 pKa = 9.52FLRR190 pKa = 11.84DD191 pKa = 3.45GLHH194 pKa = 6.18LQFKK198 pKa = 9.93GKK200 pKa = 10.13GGKK203 pKa = 6.86THH205 pKa = 7.65DD206 pKa = 4.04AGIDD210 pKa = 3.3DD211 pKa = 4.07RR212 pKa = 11.84RR213 pKa = 11.84LVKK216 pKa = 10.46IMRR219 pKa = 11.84AMHH222 pKa = 6.49EE223 pKa = 4.27LPGQRR228 pKa = 11.84LFQYY232 pKa = 10.7RR233 pKa = 11.84DD234 pKa = 3.28DD235 pKa = 4.38EE236 pKa = 5.0GNLRR240 pKa = 11.84PLDD243 pKa = 3.87SSDD246 pKa = 3.4ANDD249 pKa = 3.66YY250 pKa = 10.83LRR252 pKa = 11.84EE253 pKa = 3.98RR254 pKa = 11.84MGGPFTAKK262 pKa = 10.49DD263 pKa = 3.53FRR265 pKa = 11.84TWGATATAFRR275 pKa = 11.84LLAATPVPEE284 pKa = 5.11DD285 pKa = 3.13ATKK288 pKa = 10.62RR289 pKa = 11.84DD290 pKa = 3.35IAGVEE295 pKa = 3.71RR296 pKa = 11.84DD297 pKa = 3.76VVNAVAEE304 pKa = 4.27MLGNTPAVCRR314 pKa = 11.84KK315 pKa = 10.15SYY317 pKa = 10.75IDD319 pKa = 3.38PCVFDD324 pKa = 3.77GWQDD328 pKa = 3.43GRR330 pKa = 11.84LHH332 pKa = 6.97RR333 pKa = 11.84AAQGARR339 pKa = 11.84GPRR342 pKa = 11.84QWEE345 pKa = 4.15TATLRR350 pKa = 11.84YY351 pKa = 9.05LRR353 pKa = 11.84RR354 pKa = 11.84VKK356 pKa = 10.62KK357 pKa = 10.17PSRR360 pKa = 3.51
Molecular weight: 40.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1091155 |
22 |
2104 |
327.2 |
35.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.252 ± 0.064 | 0.824 ± 0.014 |
6.277 ± 0.036 | 5.297 ± 0.035 |
3.382 ± 0.024 | 8.723 ± 0.038 |
2.328 ± 0.022 | 4.023 ± 0.033 |
2.4 ± 0.038 | 10.559 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.245 ± 0.02 | 2.351 ± 0.028 |
5.401 ± 0.031 | 3.488 ± 0.028 |
8.098 ± 0.047 | 4.994 ± 0.027 |
4.966 ± 0.033 | 7.624 ± 0.041 |
1.55 ± 0.019 | 2.218 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
