
Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Campylobacteraceae; Campylobacter; Campylobacter jejuni; Campylobacter jejuni subsp. jejuni
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
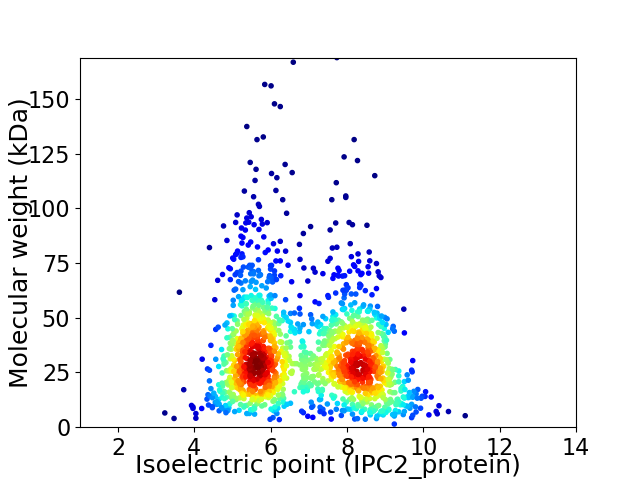
Virtual 2D-PAGE plot for 1624 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
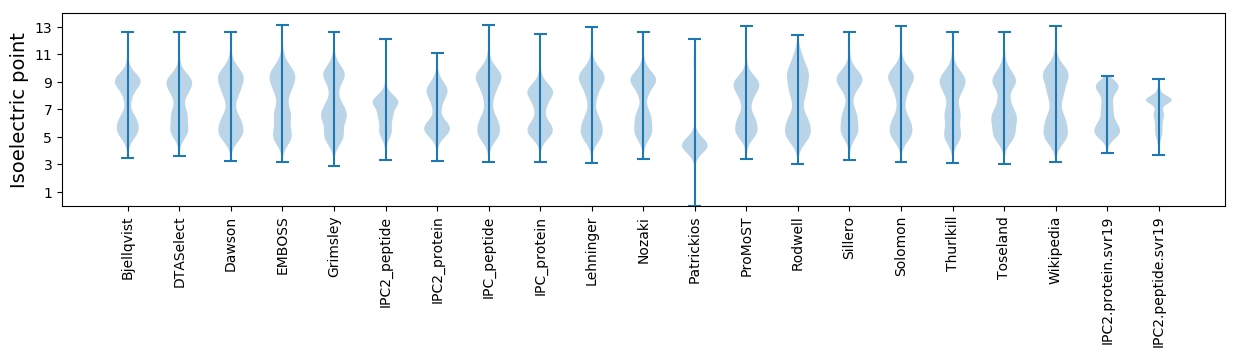
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q0P7R0|Q0P7R0_CAMJE Putative outer membrane protein OS=Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168) OX=192222 GN=Cj1721c PE=4 SV=1
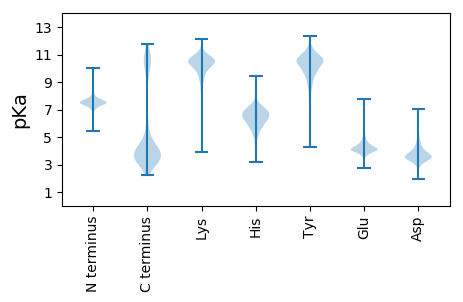
MM1 pKa = 7.68DD2 pKa = 3.72WEE4 pKa = 4.55AYY6 pKa = 10.15GYY8 pKa = 9.25EE9 pKa = 3.98IKK11 pKa = 10.85LNNYY15 pKa = 8.32MEE17 pKa = 5.4LYY19 pKa = 10.37FDD21 pKa = 4.41RR22 pKa = 11.84NGNFLGQEE30 pKa = 4.06FDD32 pKa = 3.41NN33 pKa = 4.7
MM1 pKa = 7.68DD2 pKa = 3.72WEE4 pKa = 4.55AYY6 pKa = 10.15GYY8 pKa = 9.25EE9 pKa = 3.98IKK11 pKa = 10.85LNNYY15 pKa = 8.32MEE17 pKa = 5.4LYY19 pKa = 10.37FDD21 pKa = 4.41RR22 pKa = 11.84NGNFLGQEE30 pKa = 4.06FDD32 pKa = 3.41NN33 pKa = 4.7
Molecular weight: 4.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9PPC1|RUVA_CAMJE Holliday junction ATP-dependent DNA helicase RuvA OS=Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168) OX=192222 GN=ruvA PE=3 SV=1
MM1 pKa = 7.28KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 9.91QPHH8 pKa = 5.04GTPRR12 pKa = 11.84KK13 pKa = 7.61RR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 9.07TKK25 pKa = 10.39NGRR28 pKa = 11.84KK29 pKa = 9.3VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.28KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 9.91QPHH8 pKa = 5.04GTPRR12 pKa = 11.84KK13 pKa = 7.61RR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 9.07TKK25 pKa = 10.39NGRR28 pKa = 11.84KK29 pKa = 9.3VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
508032 |
14 |
1517 |
312.8 |
35.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.81 ± 0.066 | 1.219 ± 0.025 |
5.284 ± 0.042 | 7.048 ± 0.071 |
5.998 ± 0.067 | 5.609 ± 0.063 |
1.644 ± 0.025 | 8.674 ± 0.051 |
9.507 ± 0.07 | 10.792 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.234 ± 0.023 | 6.298 ± 0.065 |
2.676 ± 0.033 | 3.129 ± 0.032 |
2.993 ± 0.035 | 6.441 ± 0.043 |
4.062 ± 0.039 | 5.255 ± 0.045 |
0.648 ± 0.017 | 3.676 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
