
Perkinsus marinus (strain ATCC 50983 / TXsc)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Perkinsozoa; Perkinsea; Perkinsida; Perkinsidae; Perkinsus; Perkinsus marinus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
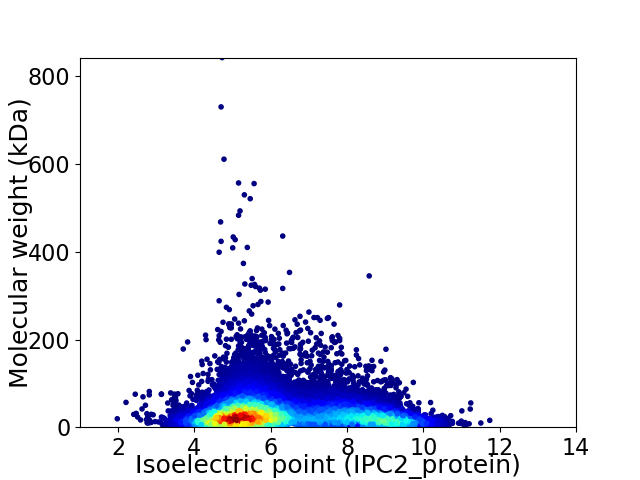
Virtual 2D-PAGE plot for 23114 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C5LP84|C5LP84_PERM5 Transposable element tc3 transposase putative OS=Perkinsus marinus (strain ATCC 50983 / TXsc) OX=423536 GN=Pmar_PMAR009612 PE=4 SV=1
MM1 pKa = 7.72SSTLDD6 pKa = 3.26YY7 pKa = 11.14SAGVNPLACDD17 pKa = 3.97YY18 pKa = 11.43DD19 pKa = 3.73WLADD23 pKa = 3.57LTLPEE28 pKa = 4.45FQCGFPEE35 pKa = 5.26GSDD38 pKa = 3.27DD39 pKa = 4.15SSWMDD44 pKa = 3.17THH46 pKa = 7.02PCDD49 pKa = 4.95SPLSPSTTASEE60 pKa = 4.49VSSLDD65 pKa = 3.25TALLIQQLLAASSSADD81 pKa = 2.76IAAAHH86 pKa = 6.32MSQCEE91 pKa = 4.02DD92 pKa = 3.72VEE94 pKa = 4.55TGATDD99 pKa = 5.18GISVRR104 pKa = 11.84KK105 pKa = 9.84VLDD108 pKa = 3.19VLIARR113 pKa = 11.84KK114 pKa = 9.7GLL116 pKa = 3.57
MM1 pKa = 7.72SSTLDD6 pKa = 3.26YY7 pKa = 11.14SAGVNPLACDD17 pKa = 3.97YY18 pKa = 11.43DD19 pKa = 3.73WLADD23 pKa = 3.57LTLPEE28 pKa = 4.45FQCGFPEE35 pKa = 5.26GSDD38 pKa = 3.27DD39 pKa = 4.15SSWMDD44 pKa = 3.17THH46 pKa = 7.02PCDD49 pKa = 4.95SPLSPSTTASEE60 pKa = 4.49VSSLDD65 pKa = 3.25TALLIQQLLAASSSADD81 pKa = 2.76IAAAHH86 pKa = 6.32MSQCEE91 pKa = 4.02DD92 pKa = 3.72VEE94 pKa = 4.55TGATDD99 pKa = 5.18GISVRR104 pKa = 11.84KK105 pKa = 9.84VLDD108 pKa = 3.19VLIARR113 pKa = 11.84KK114 pKa = 9.7GLL116 pKa = 3.57
Molecular weight: 12.2 kDa
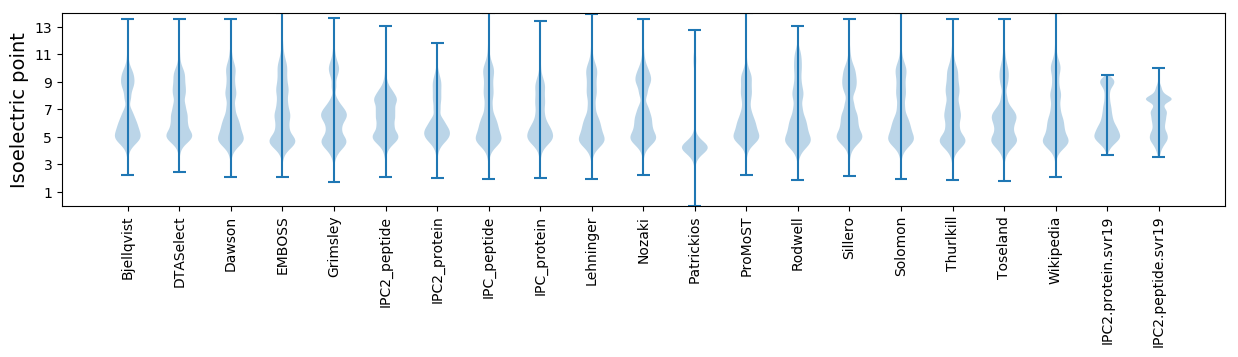
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C5KKC1|C5KKC1_PERM5 Oxidoreductase putative OS=Perkinsus marinus (strain ATCC 50983 / TXsc) OX=423536 GN=Pmar_PMAR023357 PE=4 SV=1
MM1 pKa = 7.54CCSEE5 pKa = 4.89NIGSTAARR13 pKa = 11.84TWNIRR18 pKa = 11.84PTAHH22 pKa = 6.77AGIFVIHH29 pKa = 7.19AIATTGYY36 pKa = 9.6RR37 pKa = 11.84IAEE40 pKa = 4.28IVTHH44 pKa = 7.13EE45 pKa = 3.88ITTISRR51 pKa = 11.84CCGTRR56 pKa = 11.84SGNCRR61 pKa = 11.84TRR63 pKa = 11.84SSARR67 pKa = 11.84SSSRR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84TRR75 pKa = 11.84SSARR79 pKa = 11.84SSRR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84TRR87 pKa = 11.84SSARR91 pKa = 11.84SSRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84TRR98 pKa = 11.84SSARR102 pKa = 11.84CSRR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84GRR109 pKa = 11.84TRR111 pKa = 11.84SSARR115 pKa = 11.84SSGGRR120 pKa = 11.84RR121 pKa = 11.84GRR123 pKa = 11.84TRR125 pKa = 11.84SSARR129 pKa = 11.84SSGGRR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84TRR138 pKa = 11.84SSARR142 pKa = 11.84SSGGRR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84TRR151 pKa = 11.84SSARR155 pKa = 11.84SSGGRR160 pKa = 11.84RR161 pKa = 11.84RR162 pKa = 11.84TRR164 pKa = 11.84SSARR168 pKa = 11.84CSRR171 pKa = 11.84RR172 pKa = 11.84RR173 pKa = 11.84GRR175 pKa = 11.84TRR177 pKa = 11.84SSARR181 pKa = 11.84CSRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84GRR188 pKa = 11.84TRR190 pKa = 11.84SSARR194 pKa = 11.84SSRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84GRR201 pKa = 11.84TRR203 pKa = 11.84SSARR207 pKa = 11.84CSGGRR212 pKa = 11.84RR213 pKa = 11.84RR214 pKa = 11.84TRR216 pKa = 11.84SSARR220 pKa = 11.84CSRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84GRR227 pKa = 11.84TRR229 pKa = 11.84SSARR233 pKa = 11.84SSGGRR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84TRR242 pKa = 11.84SSARR246 pKa = 11.84SSGGRR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84TRR255 pKa = 11.84SSARR259 pKa = 11.84SSGGRR264 pKa = 11.84RR265 pKa = 11.84GRR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84TRR271 pKa = 11.84SSARR275 pKa = 11.84SSGGRR280 pKa = 11.84RR281 pKa = 11.84GRR283 pKa = 11.84RR284 pKa = 11.84RR285 pKa = 11.84TRR287 pKa = 11.84SSARR291 pKa = 11.84SSGGRR296 pKa = 11.84RR297 pKa = 11.84GRR299 pKa = 11.84RR300 pKa = 11.84RR301 pKa = 11.84TRR303 pKa = 11.84RR304 pKa = 11.84AKK306 pKa = 9.55SSRR309 pKa = 11.84RR310 pKa = 11.84RR311 pKa = 11.84TRR313 pKa = 11.84RR314 pKa = 11.84RR315 pKa = 11.84TRR317 pKa = 11.84SSARR321 pKa = 11.84SSRR324 pKa = 11.84RR325 pKa = 11.84RR326 pKa = 11.84TRR328 pKa = 11.84SSARR332 pKa = 11.84SSGGRR337 pKa = 11.84RR338 pKa = 11.84GRR340 pKa = 11.84RR341 pKa = 11.84RR342 pKa = 11.84TRR344 pKa = 11.84RR345 pKa = 11.84AKK347 pKa = 9.55SSRR350 pKa = 11.84RR351 pKa = 11.84RR352 pKa = 11.84TRR354 pKa = 11.84RR355 pKa = 11.84RR356 pKa = 11.84TRR358 pKa = 11.84SSARR362 pKa = 11.84SSRR365 pKa = 11.84RR366 pKa = 11.84RR367 pKa = 11.84TRR369 pKa = 11.84SSARR373 pKa = 11.84SSARR377 pKa = 11.84NSRR380 pKa = 11.84RR381 pKa = 11.84RR382 pKa = 11.84TRR384 pKa = 11.84SSRR387 pKa = 11.84RR388 pKa = 11.84RR389 pKa = 11.84TRR391 pKa = 11.84SNRR394 pKa = 11.84RR395 pKa = 11.84RR396 pKa = 11.84TRR398 pKa = 11.84SSRR401 pKa = 11.84RR402 pKa = 11.84RR403 pKa = 11.84TRR405 pKa = 11.84SSRR408 pKa = 11.84RR409 pKa = 11.84RR410 pKa = 11.84TRR412 pKa = 11.84SSRR415 pKa = 11.84RR416 pKa = 11.84RR417 pKa = 11.84TRR419 pKa = 11.84SSRR422 pKa = 11.84RR423 pKa = 11.84RR424 pKa = 11.84SARR427 pKa = 11.84SSRR430 pKa = 11.84RR431 pKa = 11.84RR432 pKa = 11.84TRR434 pKa = 11.84RR435 pKa = 11.84RR436 pKa = 11.84TRR438 pKa = 11.84SSARR442 pKa = 11.84SSRR445 pKa = 11.84HH446 pKa = 4.01RR447 pKa = 11.84RR448 pKa = 11.84TRR450 pKa = 11.84SGMSMRR456 pKa = 11.84APGIIPLYY464 pKa = 10.56KK465 pKa = 10.35SITSFSVIPYY475 pKa = 10.56DD476 pKa = 5.2KK477 pKa = 10.19ITT479 pKa = 3.41
MM1 pKa = 7.54CCSEE5 pKa = 4.89NIGSTAARR13 pKa = 11.84TWNIRR18 pKa = 11.84PTAHH22 pKa = 6.77AGIFVIHH29 pKa = 7.19AIATTGYY36 pKa = 9.6RR37 pKa = 11.84IAEE40 pKa = 4.28IVTHH44 pKa = 7.13EE45 pKa = 3.88ITTISRR51 pKa = 11.84CCGTRR56 pKa = 11.84SGNCRR61 pKa = 11.84TRR63 pKa = 11.84SSARR67 pKa = 11.84SSSRR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84TRR75 pKa = 11.84SSARR79 pKa = 11.84SSRR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84TRR87 pKa = 11.84SSARR91 pKa = 11.84SSRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84TRR98 pKa = 11.84SSARR102 pKa = 11.84CSRR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84GRR109 pKa = 11.84TRR111 pKa = 11.84SSARR115 pKa = 11.84SSGGRR120 pKa = 11.84RR121 pKa = 11.84GRR123 pKa = 11.84TRR125 pKa = 11.84SSARR129 pKa = 11.84SSGGRR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84TRR138 pKa = 11.84SSARR142 pKa = 11.84SSGGRR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84TRR151 pKa = 11.84SSARR155 pKa = 11.84SSGGRR160 pKa = 11.84RR161 pKa = 11.84RR162 pKa = 11.84TRR164 pKa = 11.84SSARR168 pKa = 11.84CSRR171 pKa = 11.84RR172 pKa = 11.84RR173 pKa = 11.84GRR175 pKa = 11.84TRR177 pKa = 11.84SSARR181 pKa = 11.84CSRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84GRR188 pKa = 11.84TRR190 pKa = 11.84SSARR194 pKa = 11.84SSRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84GRR201 pKa = 11.84TRR203 pKa = 11.84SSARR207 pKa = 11.84CSGGRR212 pKa = 11.84RR213 pKa = 11.84RR214 pKa = 11.84TRR216 pKa = 11.84SSARR220 pKa = 11.84CSRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84GRR227 pKa = 11.84TRR229 pKa = 11.84SSARR233 pKa = 11.84SSGGRR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84TRR242 pKa = 11.84SSARR246 pKa = 11.84SSGGRR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84TRR255 pKa = 11.84SSARR259 pKa = 11.84SSGGRR264 pKa = 11.84RR265 pKa = 11.84GRR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84TRR271 pKa = 11.84SSARR275 pKa = 11.84SSGGRR280 pKa = 11.84RR281 pKa = 11.84GRR283 pKa = 11.84RR284 pKa = 11.84RR285 pKa = 11.84TRR287 pKa = 11.84SSARR291 pKa = 11.84SSGGRR296 pKa = 11.84RR297 pKa = 11.84GRR299 pKa = 11.84RR300 pKa = 11.84RR301 pKa = 11.84TRR303 pKa = 11.84RR304 pKa = 11.84AKK306 pKa = 9.55SSRR309 pKa = 11.84RR310 pKa = 11.84RR311 pKa = 11.84TRR313 pKa = 11.84RR314 pKa = 11.84RR315 pKa = 11.84TRR317 pKa = 11.84SSARR321 pKa = 11.84SSRR324 pKa = 11.84RR325 pKa = 11.84RR326 pKa = 11.84TRR328 pKa = 11.84SSARR332 pKa = 11.84SSGGRR337 pKa = 11.84RR338 pKa = 11.84GRR340 pKa = 11.84RR341 pKa = 11.84RR342 pKa = 11.84TRR344 pKa = 11.84RR345 pKa = 11.84AKK347 pKa = 9.55SSRR350 pKa = 11.84RR351 pKa = 11.84RR352 pKa = 11.84TRR354 pKa = 11.84RR355 pKa = 11.84RR356 pKa = 11.84TRR358 pKa = 11.84SSARR362 pKa = 11.84SSRR365 pKa = 11.84RR366 pKa = 11.84RR367 pKa = 11.84TRR369 pKa = 11.84SSARR373 pKa = 11.84SSARR377 pKa = 11.84NSRR380 pKa = 11.84RR381 pKa = 11.84RR382 pKa = 11.84TRR384 pKa = 11.84SSRR387 pKa = 11.84RR388 pKa = 11.84RR389 pKa = 11.84TRR391 pKa = 11.84SNRR394 pKa = 11.84RR395 pKa = 11.84RR396 pKa = 11.84TRR398 pKa = 11.84SSRR401 pKa = 11.84RR402 pKa = 11.84RR403 pKa = 11.84TRR405 pKa = 11.84SSRR408 pKa = 11.84RR409 pKa = 11.84RR410 pKa = 11.84TRR412 pKa = 11.84SSRR415 pKa = 11.84RR416 pKa = 11.84RR417 pKa = 11.84TRR419 pKa = 11.84SSRR422 pKa = 11.84RR423 pKa = 11.84RR424 pKa = 11.84SARR427 pKa = 11.84SSRR430 pKa = 11.84RR431 pKa = 11.84RR432 pKa = 11.84TRR434 pKa = 11.84RR435 pKa = 11.84RR436 pKa = 11.84TRR438 pKa = 11.84SSARR442 pKa = 11.84SSRR445 pKa = 11.84HH446 pKa = 4.01RR447 pKa = 11.84RR448 pKa = 11.84TRR450 pKa = 11.84SGMSMRR456 pKa = 11.84APGIIPLYY464 pKa = 10.56KK465 pKa = 10.35SITSFSVIPYY475 pKa = 10.56DD476 pKa = 5.2KK477 pKa = 10.19ITT479 pKa = 3.41
Molecular weight: 55.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
7493757 |
15 |
7897 |
324.2 |
35.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.411 ± 0.014 | 1.891 ± 0.009 |
5.976 ± 0.013 | 6.717 ± 0.02 |
3.383 ± 0.009 | 7.02 ± 0.016 |
2.245 ± 0.009 | 4.702 ± 0.013 |
5.054 ± 0.018 | 8.984 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.621 ± 0.008 | 3.298 ± 0.011 |
5.038 ± 0.014 | 3.507 ± 0.01 |
6.256 ± 0.018 | 7.745 ± 0.019 |
5.744 ± 0.024 | 7.285 ± 0.014 |
1.318 ± 0.007 | 2.793 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
