
Tick-associated genomovirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus ixode1
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
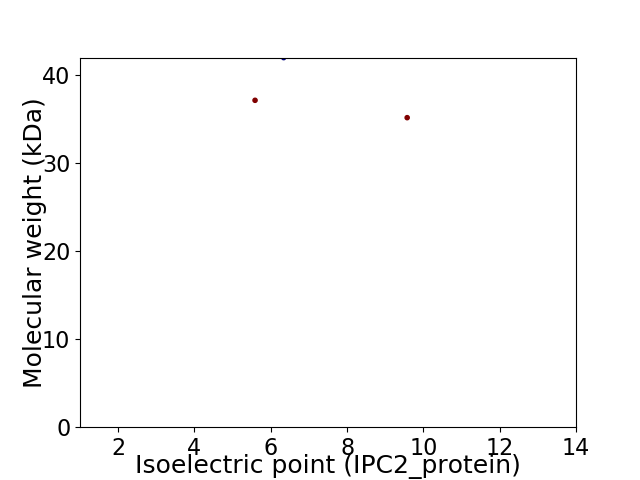
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D0Y219|A0A2D0Y219_9VIRU Capsid protein OS=Tick-associated genomovirus 3 OX=2025479 PE=4 SV=1
MM1 pKa = 7.52LFCNSKK7 pKa = 10.7YY8 pKa = 11.1VLLTYY13 pKa = 9.13AQCGDD18 pKa = 3.67LDD20 pKa = 3.59EE21 pKa = 4.86WAVSDD26 pKa = 4.61HH27 pKa = 6.68LSSLDD32 pKa = 3.46AEE34 pKa = 4.68CIVARR39 pKa = 11.84EE40 pKa = 4.05IHH42 pKa = 6.23PTTGGVHH49 pKa = 5.86LHH51 pKa = 5.79VFVDD55 pKa = 4.54FGRR58 pKa = 11.84KK59 pKa = 7.9FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84VDD66 pKa = 3.24VFDD69 pKa = 4.17VEE71 pKa = 4.89GRR73 pKa = 11.84HH74 pKa = 5.94PNVVPSKK81 pKa = 8.26GTPEE85 pKa = 3.8KK86 pKa = 10.85GYY88 pKa = 10.88DD89 pKa = 3.43YY90 pKa = 10.61AIKK93 pKa = 10.61DD94 pKa = 3.82GEE96 pKa = 4.58VVAGGLARR104 pKa = 11.84PRR106 pKa = 11.84GGRR109 pKa = 11.84PGTLSGLEE117 pKa = 4.48AIAHH121 pKa = 5.74LCEE124 pKa = 4.35TQDD127 pKa = 3.61EE128 pKa = 4.36FLDD131 pKa = 3.72IYY133 pKa = 11.48GEE135 pKa = 3.96VDD137 pKa = 2.94TRR139 pKa = 11.84GLIKK143 pKa = 10.77NFANVRR149 pKa = 11.84SYY151 pKa = 11.76AKK153 pKa = 9.1WRR155 pKa = 11.84YY156 pKa = 9.85ASTLPKK162 pKa = 10.15YY163 pKa = 9.53EE164 pKa = 4.28SPASFGEE171 pKa = 3.93FRR173 pKa = 11.84GGSDD177 pKa = 5.89GRR179 pKa = 11.84DD180 pKa = 2.8QWLAQSGIRR189 pKa = 11.84SNDD192 pKa = 2.91VVRR195 pKa = 11.84PKK197 pKa = 10.96SLVLYY202 pKa = 10.16GPSRR206 pKa = 11.84TGKK209 pKa = 8.43TSWARR214 pKa = 11.84SLGPHH219 pKa = 6.33VYY221 pKa = 10.46FGGAFSGGDD230 pKa = 3.53ALAVDD235 pKa = 5.33DD236 pKa = 4.41DD237 pKa = 4.09VKK239 pKa = 11.4YY240 pKa = 11.03AVFDD244 pKa = 4.0DD245 pKa = 3.88MRR247 pKa = 11.84GGIAFFHH254 pKa = 6.43GWKK257 pKa = 10.0DD258 pKa = 3.13WLGAQQEE265 pKa = 4.97FYY267 pKa = 10.67GQSAVSRR274 pKa = 11.84SKK276 pKa = 10.98SYY278 pKa = 11.19LSGVDD283 pKa = 3.36HH284 pKa = 6.76QFWCANRR291 pKa = 11.84DD292 pKa = 3.29PRR294 pKa = 11.84EE295 pKa = 3.89EE296 pKa = 4.18MEE298 pKa = 4.04CHH300 pKa = 6.07LFKK303 pKa = 10.88GTFCQGDD310 pKa = 4.1IDD312 pKa = 4.15WLNANCVFIEE322 pKa = 4.11VNTPIFHH329 pKa = 7.54ANRR332 pKa = 11.84EE333 pKa = 4.13
MM1 pKa = 7.52LFCNSKK7 pKa = 10.7YY8 pKa = 11.1VLLTYY13 pKa = 9.13AQCGDD18 pKa = 3.67LDD20 pKa = 3.59EE21 pKa = 4.86WAVSDD26 pKa = 4.61HH27 pKa = 6.68LSSLDD32 pKa = 3.46AEE34 pKa = 4.68CIVARR39 pKa = 11.84EE40 pKa = 4.05IHH42 pKa = 6.23PTTGGVHH49 pKa = 5.86LHH51 pKa = 5.79VFVDD55 pKa = 4.54FGRR58 pKa = 11.84KK59 pKa = 7.9FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84VDD66 pKa = 3.24VFDD69 pKa = 4.17VEE71 pKa = 4.89GRR73 pKa = 11.84HH74 pKa = 5.94PNVVPSKK81 pKa = 8.26GTPEE85 pKa = 3.8KK86 pKa = 10.85GYY88 pKa = 10.88DD89 pKa = 3.43YY90 pKa = 10.61AIKK93 pKa = 10.61DD94 pKa = 3.82GEE96 pKa = 4.58VVAGGLARR104 pKa = 11.84PRR106 pKa = 11.84GGRR109 pKa = 11.84PGTLSGLEE117 pKa = 4.48AIAHH121 pKa = 5.74LCEE124 pKa = 4.35TQDD127 pKa = 3.61EE128 pKa = 4.36FLDD131 pKa = 3.72IYY133 pKa = 11.48GEE135 pKa = 3.96VDD137 pKa = 2.94TRR139 pKa = 11.84GLIKK143 pKa = 10.77NFANVRR149 pKa = 11.84SYY151 pKa = 11.76AKK153 pKa = 9.1WRR155 pKa = 11.84YY156 pKa = 9.85ASTLPKK162 pKa = 10.15YY163 pKa = 9.53EE164 pKa = 4.28SPASFGEE171 pKa = 3.93FRR173 pKa = 11.84GGSDD177 pKa = 5.89GRR179 pKa = 11.84DD180 pKa = 2.8QWLAQSGIRR189 pKa = 11.84SNDD192 pKa = 2.91VVRR195 pKa = 11.84PKK197 pKa = 10.96SLVLYY202 pKa = 10.16GPSRR206 pKa = 11.84TGKK209 pKa = 8.43TSWARR214 pKa = 11.84SLGPHH219 pKa = 6.33VYY221 pKa = 10.46FGGAFSGGDD230 pKa = 3.53ALAVDD235 pKa = 5.33DD236 pKa = 4.41DD237 pKa = 4.09VKK239 pKa = 11.4YY240 pKa = 11.03AVFDD244 pKa = 4.0DD245 pKa = 3.88MRR247 pKa = 11.84GGIAFFHH254 pKa = 6.43GWKK257 pKa = 10.0DD258 pKa = 3.13WLGAQQEE265 pKa = 4.97FYY267 pKa = 10.67GQSAVSRR274 pKa = 11.84SKK276 pKa = 10.98SYY278 pKa = 11.19LSGVDD283 pKa = 3.36HH284 pKa = 6.76QFWCANRR291 pKa = 11.84DD292 pKa = 3.29PRR294 pKa = 11.84EE295 pKa = 3.89EE296 pKa = 4.18MEE298 pKa = 4.04CHH300 pKa = 6.07LFKK303 pKa = 10.88GTFCQGDD310 pKa = 4.1IDD312 pKa = 4.15WLNANCVFIEE322 pKa = 4.11VNTPIFHH329 pKa = 7.54ANRR332 pKa = 11.84EE333 pKa = 4.13
Molecular weight: 37.12 kDa
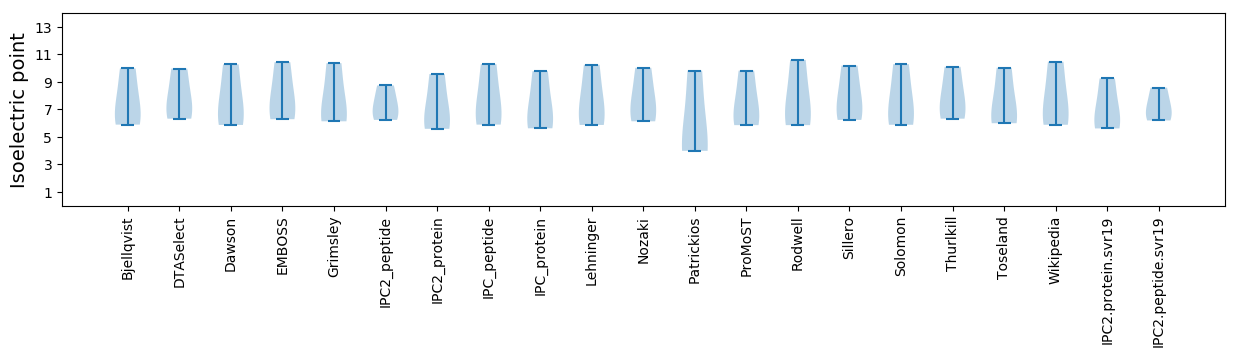
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D0Y219|A0A2D0Y219_9VIRU Capsid protein OS=Tick-associated genomovirus 3 OX=2025479 PE=4 SV=1
MM1 pKa = 7.65PRR3 pKa = 11.84FKK5 pKa = 10.85NRR7 pKa = 11.84RR8 pKa = 11.84TRR10 pKa = 11.84YY11 pKa = 7.83ARR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VGRR18 pKa = 11.84FNYY21 pKa = 9.84GNKK24 pKa = 9.4FSKK27 pKa = 10.64GKK29 pKa = 10.65GPIRR33 pKa = 11.84RR34 pKa = 11.84TKK36 pKa = 8.5RR37 pKa = 11.84TYY39 pKa = 9.79RR40 pKa = 11.84PKK42 pKa = 8.38MTKK45 pKa = 10.05KK46 pKa = 10.4RR47 pKa = 11.84ILNVTSKK54 pKa = 10.49KK55 pKa = 10.46KK56 pKa = 9.98HH57 pKa = 5.53DD58 pKa = 3.85TMLTTTNISASSQYY72 pKa = 10.82GGSAYY77 pKa = 9.42IQAPAIITGASNAIPIVWCATARR100 pKa = 11.84DD101 pKa = 3.6ASYY104 pKa = 10.87DD105 pKa = 3.5AKK107 pKa = 10.8LANPTDD113 pKa = 3.56TATRR117 pKa = 11.84TAQTCFMRR125 pKa = 11.84GLSEE129 pKa = 4.41KK130 pKa = 10.49IEE132 pKa = 4.01IQVADD137 pKa = 3.81GMPWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTYY150 pKa = 10.71RR151 pKa = 11.84NLTTLIPNTTEE162 pKa = 3.47AGKK165 pKa = 10.64YY166 pKa = 8.75FYY168 pKa = 10.82SYY170 pKa = 11.35LSIGNLGYY178 pKa = 10.9NRR180 pKa = 11.84VLNMLPHH187 pKa = 6.88PDD189 pKa = 2.83IVATLEE195 pKa = 4.12SVLFRR200 pKa = 11.84GVKK203 pKa = 10.03GKK205 pKa = 9.93DD206 pKa = 2.92WQSAMIAAVDD216 pKa = 3.82RR217 pKa = 11.84LRR219 pKa = 11.84VDD221 pKa = 3.85LKK223 pKa = 10.31YY224 pKa = 11.32DD225 pKa = 3.3KK226 pKa = 10.49TITIASGNEE235 pKa = 3.56DD236 pKa = 3.37GVIRR240 pKa = 11.84KK241 pKa = 8.44YY242 pKa = 11.15SRR244 pKa = 11.84WHH246 pKa = 5.81GMNKK250 pKa = 9.58NLVYY254 pKa = 10.79ADD256 pKa = 4.45DD257 pKa = 4.05EE258 pKa = 5.13SGGEE262 pKa = 4.09MIEE265 pKa = 4.69AYY267 pKa = 10.4HH268 pKa = 6.25SVSDD272 pKa = 3.58KK273 pKa = 11.21RR274 pKa = 11.84GMGDD278 pKa = 3.51YY279 pKa = 11.07YY280 pKa = 10.34IVDD283 pKa = 3.77YY284 pKa = 9.16FAPRR288 pKa = 11.84AGATSSNQISVGMSSTLYY306 pKa = 8.84WHH308 pKa = 7.07EE309 pKa = 4.1KK310 pKa = 9.32
MM1 pKa = 7.65PRR3 pKa = 11.84FKK5 pKa = 10.85NRR7 pKa = 11.84RR8 pKa = 11.84TRR10 pKa = 11.84YY11 pKa = 7.83ARR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VGRR18 pKa = 11.84FNYY21 pKa = 9.84GNKK24 pKa = 9.4FSKK27 pKa = 10.64GKK29 pKa = 10.65GPIRR33 pKa = 11.84RR34 pKa = 11.84TKK36 pKa = 8.5RR37 pKa = 11.84TYY39 pKa = 9.79RR40 pKa = 11.84PKK42 pKa = 8.38MTKK45 pKa = 10.05KK46 pKa = 10.4RR47 pKa = 11.84ILNVTSKK54 pKa = 10.49KK55 pKa = 10.46KK56 pKa = 9.98HH57 pKa = 5.53DD58 pKa = 3.85TMLTTTNISASSQYY72 pKa = 10.82GGSAYY77 pKa = 9.42IQAPAIITGASNAIPIVWCATARR100 pKa = 11.84DD101 pKa = 3.6ASYY104 pKa = 10.87DD105 pKa = 3.5AKK107 pKa = 10.8LANPTDD113 pKa = 3.56TATRR117 pKa = 11.84TAQTCFMRR125 pKa = 11.84GLSEE129 pKa = 4.41KK130 pKa = 10.49IEE132 pKa = 4.01IQVADD137 pKa = 3.81GMPWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTYY150 pKa = 10.71RR151 pKa = 11.84NLTTLIPNTTEE162 pKa = 3.47AGKK165 pKa = 10.64YY166 pKa = 8.75FYY168 pKa = 10.82SYY170 pKa = 11.35LSIGNLGYY178 pKa = 10.9NRR180 pKa = 11.84VLNMLPHH187 pKa = 6.88PDD189 pKa = 2.83IVATLEE195 pKa = 4.12SVLFRR200 pKa = 11.84GVKK203 pKa = 10.03GKK205 pKa = 9.93DD206 pKa = 2.92WQSAMIAAVDD216 pKa = 3.82RR217 pKa = 11.84LRR219 pKa = 11.84VDD221 pKa = 3.85LKK223 pKa = 10.31YY224 pKa = 11.32DD225 pKa = 3.3KK226 pKa = 10.49TITIASGNEE235 pKa = 3.56DD236 pKa = 3.37GVIRR240 pKa = 11.84KK241 pKa = 8.44YY242 pKa = 11.15SRR244 pKa = 11.84WHH246 pKa = 5.81GMNKK250 pKa = 9.58NLVYY254 pKa = 10.79ADD256 pKa = 4.45DD257 pKa = 4.05EE258 pKa = 5.13SGGEE262 pKa = 4.09MIEE265 pKa = 4.69AYY267 pKa = 10.4HH268 pKa = 6.25SVSDD272 pKa = 3.58KK273 pKa = 11.21RR274 pKa = 11.84GMGDD278 pKa = 3.51YY279 pKa = 11.07YY280 pKa = 10.34IVDD283 pKa = 3.77YY284 pKa = 9.16FAPRR288 pKa = 11.84AGATSSNQISVGMSSTLYY306 pKa = 8.84WHH308 pKa = 7.07EE309 pKa = 4.1KK310 pKa = 9.32
Molecular weight: 35.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1020 |
310 |
377 |
340.0 |
38.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.941 ± 0.189 | 1.961 ± 0.378 |
6.667 ± 0.587 | 4.51 ± 0.608 |
4.902 ± 0.881 | 9.804 ± 0.912 |
2.647 ± 0.393 | 4.314 ± 0.943 |
5.098 ± 0.761 | 6.667 ± 0.628 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.765 ± 0.676 | 3.824 ± 0.507 |
3.725 ± 0.076 | 2.745 ± 0.231 |
7.647 ± 0.408 | 7.353 ± 0.056 |
5.0 ± 1.298 | 6.667 ± 0.694 |
2.255 ± 0.121 | 4.51 ± 0.625 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
