
New Jersey polyomavirus-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus; Human polyomavirus 13
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
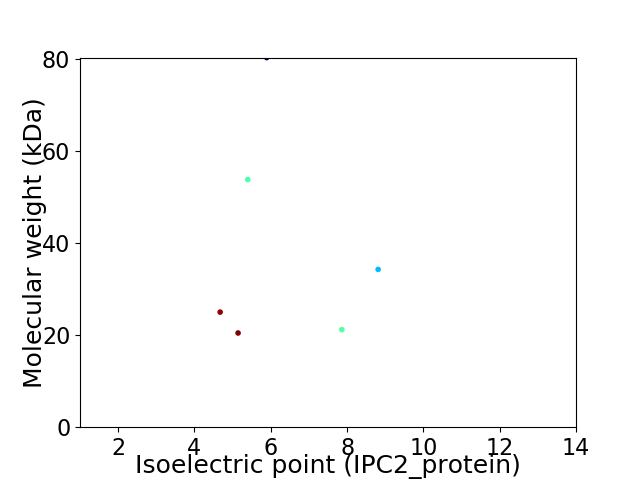
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
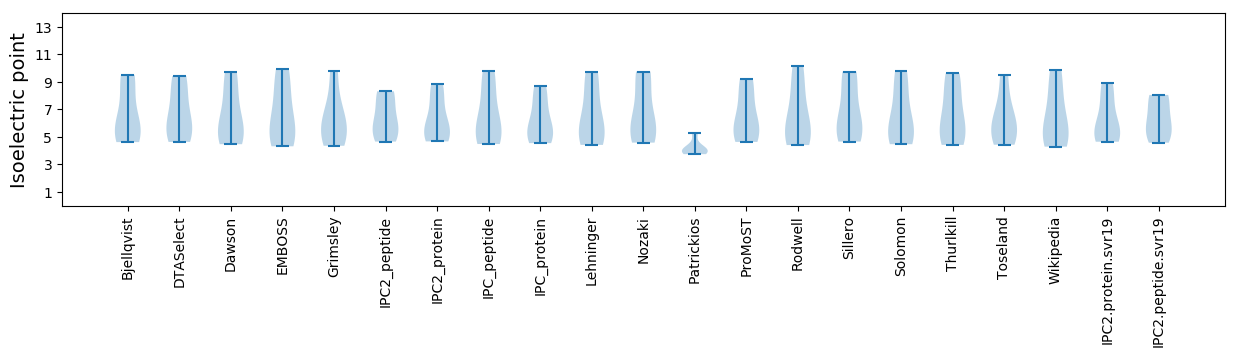
* You can choose from 21 different methods for calculating isoelectric point
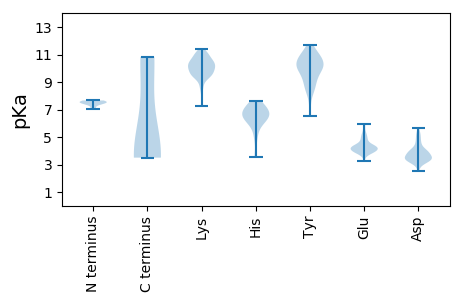
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A024B5E8|A0A024B5E8_9POLY Small T antigen OS=New Jersey polyomavirus-2013 OX=1497391 PE=4 SV=1
MM1 pKa = 7.52GGLLSSLVDD10 pKa = 4.08MIVMATEE17 pKa = 4.39LSAASGLTMEE27 pKa = 5.3ALLTGEE33 pKa = 4.6ALAALEE39 pKa = 4.41TEE41 pKa = 4.83VFSLMTVEE49 pKa = 4.22GLSGIEE55 pKa = 4.47ALAQLGWTAEE65 pKa = 4.19QFSNMAFISTTFSNAIGYY83 pKa = 8.57GVLFQTVSGISSLISAGIRR102 pKa = 11.84LGTSVSSVNRR112 pKa = 11.84HH113 pKa = 3.58QTEE116 pKa = 4.11QEE118 pKa = 3.9LEE120 pKa = 4.31TIFGKK125 pKa = 10.04IAHH128 pKa = 7.44LIHH131 pKa = 6.78VNLAFHH137 pKa = 7.1LNPFNWCEE145 pKa = 4.3SIGGTVPTEE154 pKa = 4.06FNFLSLDD161 pKa = 3.6QLSKK165 pKa = 11.01LGVVIEE171 pKa = 4.01NGRR174 pKa = 11.84WVIQTTPSFDD184 pKa = 3.41PMFEE188 pKa = 3.97SGILIEE194 pKa = 5.13RR195 pKa = 11.84YY196 pKa = 8.52EE197 pKa = 4.33PPGGAYY203 pKa = 9.28QRR205 pKa = 11.84ATPDD209 pKa = 2.54WMLPLILRR217 pKa = 11.84LNGASQEE224 pKa = 4.13KK225 pKa = 9.44TPVCPTKK232 pKa = 10.84
MM1 pKa = 7.52GGLLSSLVDD10 pKa = 4.08MIVMATEE17 pKa = 4.39LSAASGLTMEE27 pKa = 5.3ALLTGEE33 pKa = 4.6ALAALEE39 pKa = 4.41TEE41 pKa = 4.83VFSLMTVEE49 pKa = 4.22GLSGIEE55 pKa = 4.47ALAQLGWTAEE65 pKa = 4.19QFSNMAFISTTFSNAIGYY83 pKa = 8.57GVLFQTVSGISSLISAGIRR102 pKa = 11.84LGTSVSSVNRR112 pKa = 11.84HH113 pKa = 3.58QTEE116 pKa = 4.11QEE118 pKa = 3.9LEE120 pKa = 4.31TIFGKK125 pKa = 10.04IAHH128 pKa = 7.44LIHH131 pKa = 6.78VNLAFHH137 pKa = 7.1LNPFNWCEE145 pKa = 4.3SIGGTVPTEE154 pKa = 4.06FNFLSLDD161 pKa = 3.6QLSKK165 pKa = 11.01LGVVIEE171 pKa = 4.01NGRR174 pKa = 11.84WVIQTTPSFDD184 pKa = 3.41PMFEE188 pKa = 3.97SGILIEE194 pKa = 5.13RR195 pKa = 11.84YY196 pKa = 8.52EE197 pKa = 4.33PPGGAYY203 pKa = 9.28QRR205 pKa = 11.84ATPDD209 pKa = 2.54WMLPLILRR217 pKa = 11.84LNGASQEE224 pKa = 4.13KK225 pKa = 9.44TPVCPTKK232 pKa = 10.84
Molecular weight: 25.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A024B5E3|A0A024B5E3_9POLY Minor capsid protein VP2 OS=New Jersey polyomavirus-2013 OX=1497391 GN=VP2 PE=3 SV=1
MM1 pKa = 7.51EE2 pKa = 5.14KK3 pKa = 10.45VLEE6 pKa = 4.4KK7 pKa = 10.38SDD9 pKa = 4.07KK10 pKa = 10.99EE11 pKa = 3.97MLIEE15 pKa = 3.83LLGIPRR21 pKa = 11.84YY22 pKa = 10.05AYY24 pKa = 10.75GNFPIMKK31 pKa = 8.48TAYY34 pKa = 9.07KK35 pKa = 9.52RR36 pKa = 11.84ASKK39 pKa = 9.97IYY41 pKa = 10.0HH42 pKa = 6.58PDD44 pKa = 3.14KK45 pKa = 11.07GGSSEE50 pKa = 5.93KK51 pKa = 10.35MMLLNSLWQKK61 pKa = 9.68FQEE64 pKa = 4.35GLIEE68 pKa = 4.05VRR70 pKa = 11.84DD71 pKa = 3.94SEE73 pKa = 4.69VCQVSFSDD81 pKa = 5.25CYY83 pKa = 11.12DD84 pKa = 3.38SSLLKK89 pKa = 10.67CCSPKK94 pKa = 10.18VFHH97 pKa = 6.8EE98 pKa = 4.25LFLRR102 pKa = 11.84SPQCLLKK109 pKa = 11.02GPTSCSCITSCLYY122 pKa = 9.95NQHH125 pKa = 6.21RR126 pKa = 11.84QIKK129 pKa = 9.16LCGKK133 pKa = 9.4KK134 pKa = 10.02RR135 pKa = 11.84SFLIPMAAPTSEE147 pKa = 4.72RR148 pKa = 11.84DD149 pKa = 3.59MQAGAPQYY157 pKa = 11.37LPMKK161 pKa = 10.05SLTRR165 pKa = 11.84EE166 pKa = 3.64QIYY169 pKa = 8.24TVMSPPSPPPLMKK182 pKa = 10.47KK183 pKa = 8.82MKK185 pKa = 10.08PKK187 pKa = 10.63VQDD190 pKa = 3.42TTLSPLLPPQPLPPPLPPKK209 pKa = 9.99KK210 pKa = 9.74YY211 pKa = 8.83PRR213 pKa = 11.84RR214 pKa = 11.84SVSLNSPRR222 pKa = 11.84VRR224 pKa = 11.84LHH226 pKa = 6.91PEE228 pKa = 3.46ALLQEE233 pKa = 4.98EE234 pKa = 4.4IQRR237 pKa = 11.84LRR239 pKa = 11.84EE240 pKa = 3.95SLHH243 pKa = 4.73QRR245 pKa = 11.84EE246 pKa = 4.26EE247 pKa = 4.18EE248 pKa = 4.2VLKK251 pKa = 10.65IWMDD255 pKa = 4.58LILTRR260 pKa = 11.84KK261 pKa = 7.77QALRR265 pKa = 11.84AHH267 pKa = 6.16HH268 pKa = 5.66QNKK271 pKa = 9.12KK272 pKa = 9.87GKK274 pKa = 9.46VLIVLLICLLVCLILSVTLYY294 pKa = 10.97LAIKK298 pKa = 9.34QQ299 pKa = 3.69
MM1 pKa = 7.51EE2 pKa = 5.14KK3 pKa = 10.45VLEE6 pKa = 4.4KK7 pKa = 10.38SDD9 pKa = 4.07KK10 pKa = 10.99EE11 pKa = 3.97MLIEE15 pKa = 3.83LLGIPRR21 pKa = 11.84YY22 pKa = 10.05AYY24 pKa = 10.75GNFPIMKK31 pKa = 8.48TAYY34 pKa = 9.07KK35 pKa = 9.52RR36 pKa = 11.84ASKK39 pKa = 9.97IYY41 pKa = 10.0HH42 pKa = 6.58PDD44 pKa = 3.14KK45 pKa = 11.07GGSSEE50 pKa = 5.93KK51 pKa = 10.35MMLLNSLWQKK61 pKa = 9.68FQEE64 pKa = 4.35GLIEE68 pKa = 4.05VRR70 pKa = 11.84DD71 pKa = 3.94SEE73 pKa = 4.69VCQVSFSDD81 pKa = 5.25CYY83 pKa = 11.12DD84 pKa = 3.38SSLLKK89 pKa = 10.67CCSPKK94 pKa = 10.18VFHH97 pKa = 6.8EE98 pKa = 4.25LFLRR102 pKa = 11.84SPQCLLKK109 pKa = 11.02GPTSCSCITSCLYY122 pKa = 9.95NQHH125 pKa = 6.21RR126 pKa = 11.84QIKK129 pKa = 9.16LCGKK133 pKa = 9.4KK134 pKa = 10.02RR135 pKa = 11.84SFLIPMAAPTSEE147 pKa = 4.72RR148 pKa = 11.84DD149 pKa = 3.59MQAGAPQYY157 pKa = 11.37LPMKK161 pKa = 10.05SLTRR165 pKa = 11.84EE166 pKa = 3.64QIYY169 pKa = 8.24TVMSPPSPPPLMKK182 pKa = 10.47KK183 pKa = 8.82MKK185 pKa = 10.08PKK187 pKa = 10.63VQDD190 pKa = 3.42TTLSPLLPPQPLPPPLPPKK209 pKa = 9.99KK210 pKa = 9.74YY211 pKa = 8.83PRR213 pKa = 11.84RR214 pKa = 11.84SVSLNSPRR222 pKa = 11.84VRR224 pKa = 11.84LHH226 pKa = 6.91PEE228 pKa = 3.46ALLQEE233 pKa = 4.98EE234 pKa = 4.4IQRR237 pKa = 11.84LRR239 pKa = 11.84EE240 pKa = 3.95SLHH243 pKa = 4.73QRR245 pKa = 11.84EE246 pKa = 4.26EE247 pKa = 4.18EE248 pKa = 4.2VLKK251 pKa = 10.65IWMDD255 pKa = 4.58LILTRR260 pKa = 11.84KK261 pKa = 7.77QALRR265 pKa = 11.84AHH267 pKa = 6.16HH268 pKa = 5.66QNKK271 pKa = 9.12KK272 pKa = 9.87GKK274 pKa = 9.46VLIVLLICLLVCLILSVTLYY294 pKa = 10.97LAIKK298 pKa = 9.34QQ299 pKa = 3.69
Molecular weight: 34.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2101 |
183 |
711 |
350.2 |
39.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.807 ± 0.546 | 2.713 ± 0.667 |
4.046 ± 0.841 | 6.901 ± 0.248 |
4.522 ± 0.879 | 5.997 ± 0.935 |
1.904 ± 0.137 | 5.045 ± 0.651 |
6.901 ± 1.071 | 10.9 ± 0.932 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.999 ± 0.309 | 3.808 ± 0.542 |
6.711 ± 1.101 | 4.522 ± 0.245 |
3.76 ± 0.345 | 8.425 ± 0.767 |
5.474 ± 0.668 | 5.474 ± 0.403 |
1.285 ± 0.308 | 2.808 ± 0.348 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
