
Circoviridae TaCV1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.59
Get precalculated fractions of proteins
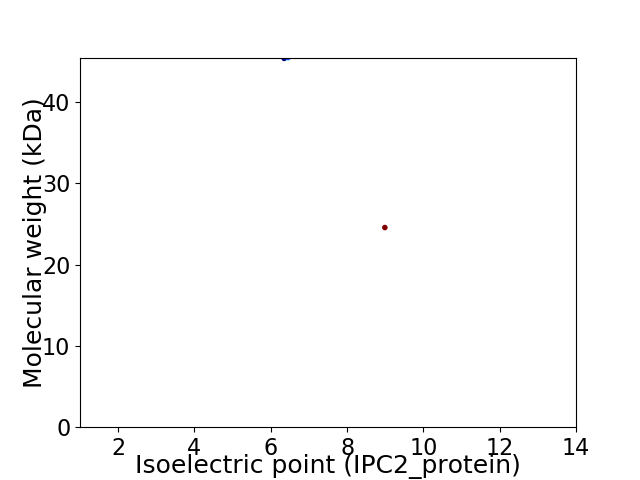
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L2P5L4|A0A2L2P5L4_9CIRC Putative viral capsid OS=Circoviridae TaCV1 OX=2094724 PE=4 SV=1
MM1 pKa = 7.64SEE3 pKa = 4.08KK4 pKa = 10.66ASNNSKK10 pKa = 9.93SRR12 pKa = 11.84NYY14 pKa = 10.15VFTIFNVGEE23 pKa = 4.33DD24 pKa = 3.67EE25 pKa = 4.63SDD27 pKa = 3.44VLGRR31 pKa = 11.84LYY33 pKa = 11.01EE34 pKa = 4.2EE35 pKa = 5.16GFCNNIFFTHH45 pKa = 6.25EE46 pKa = 3.8LGEE49 pKa = 4.37NKK51 pKa = 7.64EE52 pKa = 4.47TPHH55 pKa = 6.18IQGFLSCSSSRR66 pKa = 11.84KK67 pKa = 9.8LLTVKK72 pKa = 10.82NKK74 pKa = 10.02INSLFEE80 pKa = 4.01NAPNPHH86 pKa = 5.43VEE88 pKa = 4.08KK89 pKa = 10.92ANGSAYY95 pKa = 10.37QNYY98 pKa = 9.91IYY100 pKa = 9.19ITKK103 pKa = 9.5EE104 pKa = 3.72LKK106 pKa = 10.2EE107 pKa = 4.13NPLLKK112 pKa = 10.35HH113 pKa = 6.62RR114 pKa = 11.84IYY116 pKa = 11.09GNRR119 pKa = 11.84PLPPGKK125 pKa = 10.27KK126 pKa = 9.15RR127 pKa = 11.84DD128 pKa = 3.63DD129 pKa = 3.99SKK131 pKa = 11.58FEE133 pKa = 4.44SYY135 pKa = 11.6VNLLEE140 pKa = 4.74KK141 pKa = 11.27GEE143 pKa = 3.87IRR145 pKa = 11.84LRR147 pKa = 11.84DD148 pKa = 3.55IEE150 pKa = 4.73RR151 pKa = 11.84EE152 pKa = 4.03DD153 pKa = 3.29LAHH156 pKa = 5.44YY157 pKa = 9.34QRR159 pKa = 11.84HH160 pKa = 4.51EE161 pKa = 4.0SFYY164 pKa = 10.85LGVYY168 pKa = 9.26SKK170 pKa = 11.34LMRR173 pKa = 11.84RR174 pKa = 11.84GIKK177 pKa = 10.07PPMFVSWFSGPSGSGKK193 pKa = 10.44SFTAEE198 pKa = 4.36AIAEE202 pKa = 4.02ALNFDD207 pKa = 4.18IYY209 pKa = 11.24EE210 pKa = 4.32VTCDD214 pKa = 3.17NGFFGNYY221 pKa = 9.25GGEE224 pKa = 4.9EE225 pKa = 3.65LALWNDD231 pKa = 3.25YY232 pKa = 10.76RR233 pKa = 11.84AGCVSFHH240 pKa = 5.97TLLRR244 pKa = 11.84ITDD247 pKa = 3.86RR248 pKa = 11.84KK249 pKa = 10.84GSMINVKK256 pKa = 9.7GSKK259 pKa = 10.17VFFCPRR265 pKa = 11.84IQIFTSPDD273 pKa = 3.34GIEE276 pKa = 4.11SVKK279 pKa = 9.78TYY281 pKa = 10.18EE282 pKa = 3.84MEE284 pKa = 4.05YY285 pKa = 9.7MNSHH289 pKa = 5.85TRR291 pKa = 11.84RR292 pKa = 11.84LDD294 pKa = 3.33YY295 pKa = 10.75KK296 pKa = 10.29FQQLLRR302 pKa = 11.84RR303 pKa = 11.84VSYY306 pKa = 11.14VVDD309 pKa = 4.88FKK311 pKa = 11.55LSNDD315 pKa = 3.59DD316 pKa = 3.88SVPTLKK322 pKa = 10.23EE323 pKa = 3.67VKK325 pKa = 9.81RR326 pKa = 11.84NSEE329 pKa = 4.09EE330 pKa = 3.95VVKK333 pKa = 10.83RR334 pKa = 11.84FLGVYY339 pKa = 9.93KK340 pKa = 10.7YY341 pKa = 10.48FLIDD345 pKa = 3.51NGFEE349 pKa = 4.21EE350 pKa = 4.91YY351 pKa = 11.01VKK353 pKa = 10.7LLPALTDD360 pKa = 3.41VEE362 pKa = 4.56PIKK365 pKa = 11.02INTKK369 pKa = 9.17LVKK372 pKa = 10.17EE373 pKa = 4.16DD374 pKa = 4.47EE375 pKa = 4.33EE376 pKa = 5.11CKK378 pKa = 10.9VYY380 pKa = 10.13TVDD383 pKa = 3.64NAVSGYY389 pKa = 8.8EE390 pKa = 3.84HH391 pKa = 7.38ALL393 pKa = 3.52
MM1 pKa = 7.64SEE3 pKa = 4.08KK4 pKa = 10.66ASNNSKK10 pKa = 9.93SRR12 pKa = 11.84NYY14 pKa = 10.15VFTIFNVGEE23 pKa = 4.33DD24 pKa = 3.67EE25 pKa = 4.63SDD27 pKa = 3.44VLGRR31 pKa = 11.84LYY33 pKa = 11.01EE34 pKa = 4.2EE35 pKa = 5.16GFCNNIFFTHH45 pKa = 6.25EE46 pKa = 3.8LGEE49 pKa = 4.37NKK51 pKa = 7.64EE52 pKa = 4.47TPHH55 pKa = 6.18IQGFLSCSSSRR66 pKa = 11.84KK67 pKa = 9.8LLTVKK72 pKa = 10.82NKK74 pKa = 10.02INSLFEE80 pKa = 4.01NAPNPHH86 pKa = 5.43VEE88 pKa = 4.08KK89 pKa = 10.92ANGSAYY95 pKa = 10.37QNYY98 pKa = 9.91IYY100 pKa = 9.19ITKK103 pKa = 9.5EE104 pKa = 3.72LKK106 pKa = 10.2EE107 pKa = 4.13NPLLKK112 pKa = 10.35HH113 pKa = 6.62RR114 pKa = 11.84IYY116 pKa = 11.09GNRR119 pKa = 11.84PLPPGKK125 pKa = 10.27KK126 pKa = 9.15RR127 pKa = 11.84DD128 pKa = 3.63DD129 pKa = 3.99SKK131 pKa = 11.58FEE133 pKa = 4.44SYY135 pKa = 11.6VNLLEE140 pKa = 4.74KK141 pKa = 11.27GEE143 pKa = 3.87IRR145 pKa = 11.84LRR147 pKa = 11.84DD148 pKa = 3.55IEE150 pKa = 4.73RR151 pKa = 11.84EE152 pKa = 4.03DD153 pKa = 3.29LAHH156 pKa = 5.44YY157 pKa = 9.34QRR159 pKa = 11.84HH160 pKa = 4.51EE161 pKa = 4.0SFYY164 pKa = 10.85LGVYY168 pKa = 9.26SKK170 pKa = 11.34LMRR173 pKa = 11.84RR174 pKa = 11.84GIKK177 pKa = 10.07PPMFVSWFSGPSGSGKK193 pKa = 10.44SFTAEE198 pKa = 4.36AIAEE202 pKa = 4.02ALNFDD207 pKa = 4.18IYY209 pKa = 11.24EE210 pKa = 4.32VTCDD214 pKa = 3.17NGFFGNYY221 pKa = 9.25GGEE224 pKa = 4.9EE225 pKa = 3.65LALWNDD231 pKa = 3.25YY232 pKa = 10.76RR233 pKa = 11.84AGCVSFHH240 pKa = 5.97TLLRR244 pKa = 11.84ITDD247 pKa = 3.86RR248 pKa = 11.84KK249 pKa = 10.84GSMINVKK256 pKa = 9.7GSKK259 pKa = 10.17VFFCPRR265 pKa = 11.84IQIFTSPDD273 pKa = 3.34GIEE276 pKa = 4.11SVKK279 pKa = 9.78TYY281 pKa = 10.18EE282 pKa = 3.84MEE284 pKa = 4.05YY285 pKa = 9.7MNSHH289 pKa = 5.85TRR291 pKa = 11.84RR292 pKa = 11.84LDD294 pKa = 3.33YY295 pKa = 10.75KK296 pKa = 10.29FQQLLRR302 pKa = 11.84RR303 pKa = 11.84VSYY306 pKa = 11.14VVDD309 pKa = 4.88FKK311 pKa = 11.55LSNDD315 pKa = 3.59DD316 pKa = 3.88SVPTLKK322 pKa = 10.23EE323 pKa = 3.67VKK325 pKa = 9.81RR326 pKa = 11.84NSEE329 pKa = 4.09EE330 pKa = 3.95VVKK333 pKa = 10.83RR334 pKa = 11.84FLGVYY339 pKa = 9.93KK340 pKa = 10.7YY341 pKa = 10.48FLIDD345 pKa = 3.51NGFEE349 pKa = 4.21EE350 pKa = 4.91YY351 pKa = 11.01VKK353 pKa = 10.7LLPALTDD360 pKa = 3.41VEE362 pKa = 4.56PIKK365 pKa = 11.02INTKK369 pKa = 9.17LVKK372 pKa = 10.17EE373 pKa = 4.16DD374 pKa = 4.47EE375 pKa = 4.33EE376 pKa = 5.11CKK378 pKa = 10.9VYY380 pKa = 10.13TVDD383 pKa = 3.64NAVSGYY389 pKa = 8.8EE390 pKa = 3.84HH391 pKa = 7.38ALL393 pKa = 3.52
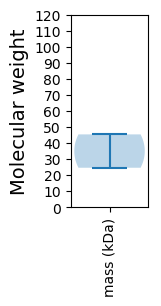
Molecular weight: 45.36 kDa
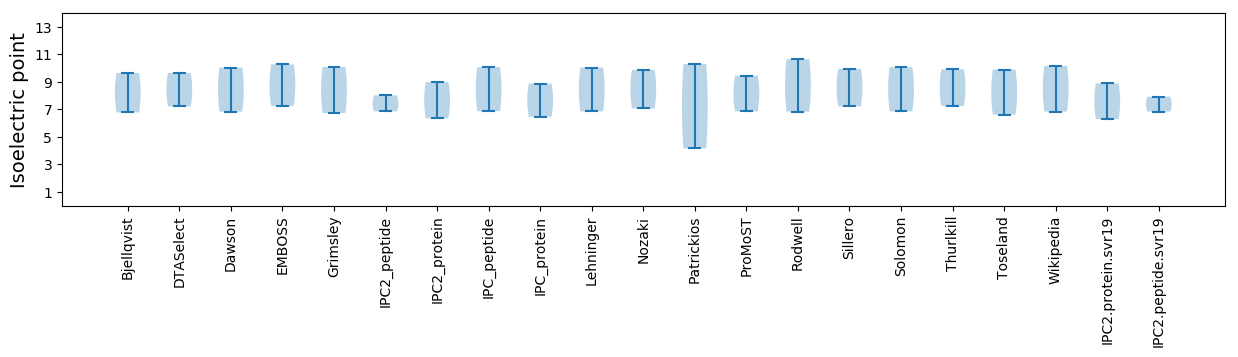
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L2P5L4|A0A2L2P5L4_9CIRC Putative viral capsid OS=Circoviridae TaCV1 OX=2094724 PE=4 SV=1
MM1 pKa = 7.63SKK3 pKa = 9.95RR4 pKa = 11.84HH5 pKa = 4.94TRR7 pKa = 11.84SYY9 pKa = 10.19LQEE12 pKa = 3.71INTFKK17 pKa = 11.0KK18 pKa = 9.86KK19 pKa = 10.35VKK21 pKa = 10.53AIDD24 pKa = 3.47RR25 pKa = 11.84NEE27 pKa = 3.85IPSYY31 pKa = 9.3TDD33 pKa = 2.52IMHH36 pKa = 6.35MRR38 pKa = 11.84NIQLALEE45 pKa = 4.1FLTINANPEE54 pKa = 3.91KK55 pKa = 10.85AIVTQKK61 pKa = 11.23AFAKK65 pKa = 9.27SKK67 pKa = 11.04NMGTATLRR75 pKa = 11.84KK76 pKa = 8.75AIKK79 pKa = 9.82EE80 pKa = 3.97ITHH83 pKa = 5.39SGMRR87 pKa = 11.84KK88 pKa = 9.48VNPKK92 pKa = 10.18SNNIQKK98 pKa = 9.47YY99 pKa = 9.48QNDD102 pKa = 3.97KK103 pKa = 11.45KK104 pKa = 11.1EE105 pKa = 4.13LLQKK109 pKa = 8.61TRR111 pKa = 11.84EE112 pKa = 4.07YY113 pKa = 11.24GEE115 pKa = 3.87QALTQEE121 pKa = 4.36EE122 pKa = 4.44QLRR125 pKa = 11.84LQQIQEE131 pKa = 4.22KK132 pKa = 9.07EE133 pKa = 3.85LQRR136 pKa = 11.84IEE138 pKa = 4.15KK139 pKa = 10.04CKK141 pKa = 9.37KK142 pKa = 8.18TKK144 pKa = 9.73TKK146 pKa = 10.92AKK148 pKa = 10.34AKK150 pKa = 9.1HH151 pKa = 5.77TDD153 pKa = 2.74NEE155 pKa = 4.5YY156 pKa = 10.63EE157 pKa = 3.89VRR159 pKa = 11.84GGYY162 pKa = 9.13RR163 pKa = 11.84EE164 pKa = 4.3HH165 pKa = 6.95EE166 pKa = 4.07NDD168 pKa = 3.81YY169 pKa = 11.41EE170 pKa = 4.13EE171 pKa = 5.29QEE173 pKa = 4.36EE174 pKa = 4.25NTIKK178 pKa = 10.52HH179 pKa = 5.81RR180 pKa = 11.84NINRR184 pKa = 11.84ALQTLINSKK193 pKa = 9.19NEE195 pKa = 3.81KK196 pKa = 9.23EE197 pKa = 4.06QLRR200 pKa = 11.84QKK202 pKa = 9.93LTIDD206 pKa = 3.48EE207 pKa = 4.37
MM1 pKa = 7.63SKK3 pKa = 9.95RR4 pKa = 11.84HH5 pKa = 4.94TRR7 pKa = 11.84SYY9 pKa = 10.19LQEE12 pKa = 3.71INTFKK17 pKa = 11.0KK18 pKa = 9.86KK19 pKa = 10.35VKK21 pKa = 10.53AIDD24 pKa = 3.47RR25 pKa = 11.84NEE27 pKa = 3.85IPSYY31 pKa = 9.3TDD33 pKa = 2.52IMHH36 pKa = 6.35MRR38 pKa = 11.84NIQLALEE45 pKa = 4.1FLTINANPEE54 pKa = 3.91KK55 pKa = 10.85AIVTQKK61 pKa = 11.23AFAKK65 pKa = 9.27SKK67 pKa = 11.04NMGTATLRR75 pKa = 11.84KK76 pKa = 8.75AIKK79 pKa = 9.82EE80 pKa = 3.97ITHH83 pKa = 5.39SGMRR87 pKa = 11.84KK88 pKa = 9.48VNPKK92 pKa = 10.18SNNIQKK98 pKa = 9.47YY99 pKa = 9.48QNDD102 pKa = 3.97KK103 pKa = 11.45KK104 pKa = 11.1EE105 pKa = 4.13LLQKK109 pKa = 8.61TRR111 pKa = 11.84EE112 pKa = 4.07YY113 pKa = 11.24GEE115 pKa = 3.87QALTQEE121 pKa = 4.36EE122 pKa = 4.44QLRR125 pKa = 11.84LQQIQEE131 pKa = 4.22KK132 pKa = 9.07EE133 pKa = 3.85LQRR136 pKa = 11.84IEE138 pKa = 4.15KK139 pKa = 10.04CKK141 pKa = 9.37KK142 pKa = 8.18TKK144 pKa = 9.73TKK146 pKa = 10.92AKK148 pKa = 10.34AKK150 pKa = 9.1HH151 pKa = 5.77TDD153 pKa = 2.74NEE155 pKa = 4.5YY156 pKa = 10.63EE157 pKa = 3.89VRR159 pKa = 11.84GGYY162 pKa = 9.13RR163 pKa = 11.84EE164 pKa = 4.3HH165 pKa = 6.95EE166 pKa = 4.07NDD168 pKa = 3.81YY169 pKa = 11.41EE170 pKa = 4.13EE171 pKa = 5.29QEE173 pKa = 4.36EE174 pKa = 4.25NTIKK178 pKa = 10.52HH179 pKa = 5.81RR180 pKa = 11.84NINRR184 pKa = 11.84ALQTLINSKK193 pKa = 9.19NEE195 pKa = 3.81KK196 pKa = 9.23EE197 pKa = 4.06QLRR200 pKa = 11.84QKK202 pKa = 9.93LTIDD206 pKa = 3.48EE207 pKa = 4.37
Molecular weight: 24.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
600 |
207 |
393 |
300.0 |
34.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.333 ± 0.814 | 1.167 ± 0.38 |
4.167 ± 0.705 | 10.0 ± 0.886 |
4.5 ± 1.696 | 5.0 ± 1.437 |
2.5 ± 0.222 | 6.0 ± 0.962 |
10.167 ± 2.137 | 8.167 ± 0.512 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.833 ± 0.324 | 7.333 ± 0.489 |
3.0 ± 0.862 | 3.833 ± 2.435 |
6.0 ± 0.424 | 6.333 ± 1.641 |
5.333 ± 1.332 | 5.167 ± 1.798 |
0.333 ± 0.185 | 4.833 ± 0.807 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
