
Sulfurimonas autotrophica (strain ATCC BAA-671 / DSM 16294 / JCM 11897 / OK10)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Thiovulaceae; Sulfurimonas; Sulfurimonas autotrophica
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
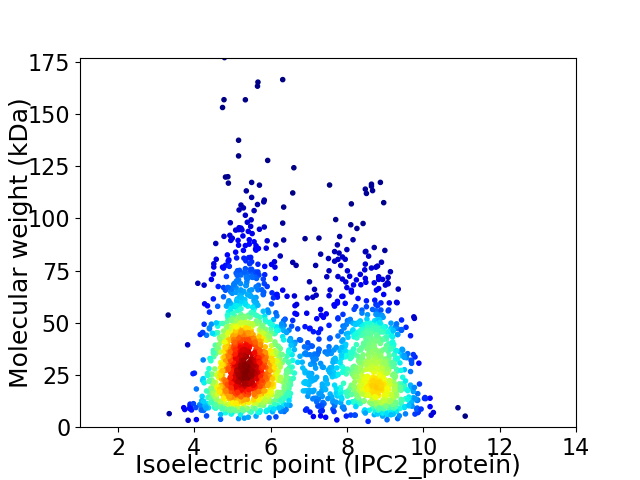
Virtual 2D-PAGE plot for 2155 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E0UPK6|E0UPK6_SULAO HI0933 family protein OS=Sulfurimonas autotrophica (strain ATCC BAA-671 / DSM 16294 / JCM 11897 / OK10) OX=563040 GN=Saut_0549 PE=4 SV=1
MM1 pKa = 7.8LSTILEE7 pKa = 4.16SLYY10 pKa = 11.15LKK12 pKa = 10.61VLVNIVVQRR21 pKa = 11.84EE22 pKa = 4.16STLVHH27 pKa = 6.84IEE29 pKa = 3.97LCSKK33 pKa = 10.48KK34 pKa = 10.81GIVDD38 pKa = 3.68EE39 pKa = 4.86VSTEE43 pKa = 4.1FNTTLLNNQMYY54 pKa = 10.53DD55 pKa = 4.12FISEE59 pKa = 4.4YY60 pKa = 10.25IKK62 pKa = 10.32EE63 pKa = 4.2SPYY66 pKa = 10.49FYY68 pKa = 10.54ISILDD73 pKa = 3.56TSIVQGAIPTCEE85 pKa = 4.03KK86 pKa = 11.02NKK88 pKa = 10.34LSYY91 pKa = 11.06YY92 pKa = 9.9YY93 pKa = 9.84DD94 pKa = 3.75LSMSEE99 pKa = 4.43YY100 pKa = 10.88KK101 pKa = 11.01CFDD104 pKa = 3.82NKK106 pKa = 10.08WNFYY110 pKa = 6.47TAKK113 pKa = 10.58NDD115 pKa = 2.63IHH117 pKa = 7.64QIEE120 pKa = 4.79RR121 pKa = 11.84EE122 pKa = 4.08YY123 pKa = 11.24EE124 pKa = 4.15DD125 pKa = 4.07IGIDD129 pKa = 3.51MIFSPFVILANFFQDD144 pKa = 4.65KK145 pKa = 10.46INNNLAMYY153 pKa = 10.84ALVQDD158 pKa = 4.29GFISVAVFEE167 pKa = 4.5NSQLLYY173 pKa = 10.91AEE175 pKa = 4.84HH176 pKa = 7.88LDD178 pKa = 3.61MLTGDD183 pKa = 3.94EE184 pKa = 5.11NEE186 pKa = 5.44DD187 pKa = 3.15ILLNEE192 pKa = 4.92NIEE195 pKa = 4.1EE196 pKa = 4.69DD197 pKa = 3.87LDD199 pKa = 4.34LDD201 pKa = 4.54SGIDD205 pKa = 3.98LEE207 pKa = 6.32DD208 pKa = 4.23IDD210 pKa = 5.62VDD212 pKa = 3.74EE213 pKa = 5.59GDD215 pKa = 4.05INSLDD220 pKa = 3.63EE221 pKa = 4.89FGDD224 pKa = 3.92IEE226 pKa = 5.35DD227 pKa = 4.9LDD229 pKa = 4.03SLEE232 pKa = 5.79DD233 pKa = 3.48IDD235 pKa = 5.45EE236 pKa = 4.67FSDD239 pKa = 3.5NKK241 pKa = 10.78DD242 pKa = 3.43VEE244 pKa = 4.45EE245 pKa = 4.28EE246 pKa = 4.46LYY248 pKa = 10.86EE249 pKa = 4.17SDD251 pKa = 5.1KK252 pKa = 11.08ILPEE256 pKa = 4.32DD257 pKa = 4.08EE258 pKa = 4.09EE259 pKa = 5.18NEE261 pKa = 4.18AFNEE265 pKa = 4.18DD266 pKa = 3.7YY267 pKa = 11.08QRR269 pKa = 11.84FSLMQSAVANFYY281 pKa = 10.8RR282 pKa = 11.84DD283 pKa = 3.3DD284 pKa = 3.9KK285 pKa = 11.05YY286 pKa = 11.28EE287 pKa = 3.91SRR289 pKa = 11.84FIEE292 pKa = 3.91NVYY295 pKa = 10.35IADD298 pKa = 4.0CAGVTNDD305 pKa = 3.17LKK307 pKa = 11.12KK308 pKa = 10.8YY309 pKa = 10.39LEE311 pKa = 4.12EE312 pKa = 3.91EE313 pKa = 4.13MFFNVYY319 pKa = 7.61VRR321 pKa = 11.84RR322 pKa = 11.84ADD324 pKa = 3.69LGVEE328 pKa = 4.11VCEE331 pKa = 4.01LAKK334 pKa = 10.23MEE336 pKa = 5.07LGLL339 pKa = 4.78
MM1 pKa = 7.8LSTILEE7 pKa = 4.16SLYY10 pKa = 11.15LKK12 pKa = 10.61VLVNIVVQRR21 pKa = 11.84EE22 pKa = 4.16STLVHH27 pKa = 6.84IEE29 pKa = 3.97LCSKK33 pKa = 10.48KK34 pKa = 10.81GIVDD38 pKa = 3.68EE39 pKa = 4.86VSTEE43 pKa = 4.1FNTTLLNNQMYY54 pKa = 10.53DD55 pKa = 4.12FISEE59 pKa = 4.4YY60 pKa = 10.25IKK62 pKa = 10.32EE63 pKa = 4.2SPYY66 pKa = 10.49FYY68 pKa = 10.54ISILDD73 pKa = 3.56TSIVQGAIPTCEE85 pKa = 4.03KK86 pKa = 11.02NKK88 pKa = 10.34LSYY91 pKa = 11.06YY92 pKa = 9.9YY93 pKa = 9.84DD94 pKa = 3.75LSMSEE99 pKa = 4.43YY100 pKa = 10.88KK101 pKa = 11.01CFDD104 pKa = 3.82NKK106 pKa = 10.08WNFYY110 pKa = 6.47TAKK113 pKa = 10.58NDD115 pKa = 2.63IHH117 pKa = 7.64QIEE120 pKa = 4.79RR121 pKa = 11.84EE122 pKa = 4.08YY123 pKa = 11.24EE124 pKa = 4.15DD125 pKa = 4.07IGIDD129 pKa = 3.51MIFSPFVILANFFQDD144 pKa = 4.65KK145 pKa = 10.46INNNLAMYY153 pKa = 10.84ALVQDD158 pKa = 4.29GFISVAVFEE167 pKa = 4.5NSQLLYY173 pKa = 10.91AEE175 pKa = 4.84HH176 pKa = 7.88LDD178 pKa = 3.61MLTGDD183 pKa = 3.94EE184 pKa = 5.11NEE186 pKa = 5.44DD187 pKa = 3.15ILLNEE192 pKa = 4.92NIEE195 pKa = 4.1EE196 pKa = 4.69DD197 pKa = 3.87LDD199 pKa = 4.34LDD201 pKa = 4.54SGIDD205 pKa = 3.98LEE207 pKa = 6.32DD208 pKa = 4.23IDD210 pKa = 5.62VDD212 pKa = 3.74EE213 pKa = 5.59GDD215 pKa = 4.05INSLDD220 pKa = 3.63EE221 pKa = 4.89FGDD224 pKa = 3.92IEE226 pKa = 5.35DD227 pKa = 4.9LDD229 pKa = 4.03SLEE232 pKa = 5.79DD233 pKa = 3.48IDD235 pKa = 5.45EE236 pKa = 4.67FSDD239 pKa = 3.5NKK241 pKa = 10.78DD242 pKa = 3.43VEE244 pKa = 4.45EE245 pKa = 4.28EE246 pKa = 4.46LYY248 pKa = 10.86EE249 pKa = 4.17SDD251 pKa = 5.1KK252 pKa = 11.08ILPEE256 pKa = 4.32DD257 pKa = 4.08EE258 pKa = 4.09EE259 pKa = 5.18NEE261 pKa = 4.18AFNEE265 pKa = 4.18DD266 pKa = 3.7YY267 pKa = 11.08QRR269 pKa = 11.84FSLMQSAVANFYY281 pKa = 10.8RR282 pKa = 11.84DD283 pKa = 3.3DD284 pKa = 3.9KK285 pKa = 11.05YY286 pKa = 11.28EE287 pKa = 3.91SRR289 pKa = 11.84FIEE292 pKa = 3.91NVYY295 pKa = 10.35IADD298 pKa = 4.0CAGVTNDD305 pKa = 3.17LKK307 pKa = 11.12KK308 pKa = 10.8YY309 pKa = 10.39LEE311 pKa = 4.12EE312 pKa = 3.91EE313 pKa = 4.13MFFNVYY319 pKa = 7.61VRR321 pKa = 11.84RR322 pKa = 11.84ADD324 pKa = 3.69LGVEE328 pKa = 4.11VCEE331 pKa = 4.01LAKK334 pKa = 10.23MEE336 pKa = 5.07LGLL339 pKa = 4.78
Molecular weight: 39.44 kDa
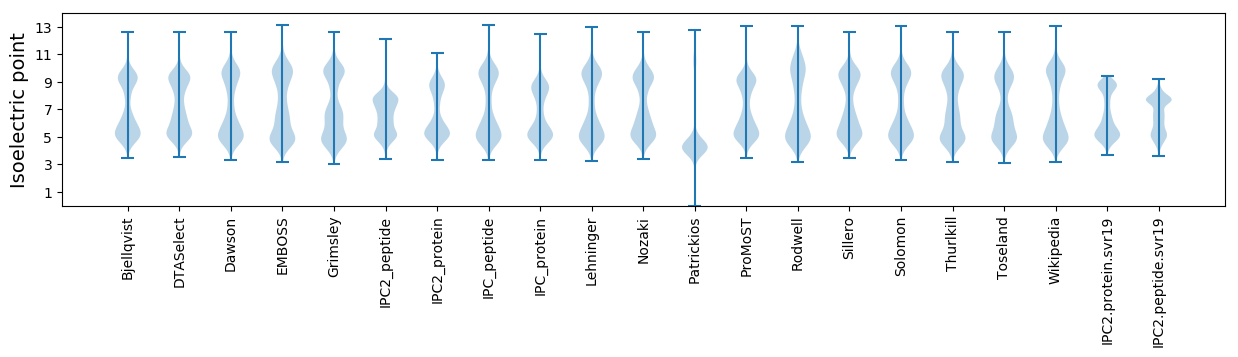
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E0UPP8|E0UPP8_SULAO Peptidase M23 OS=Sulfurimonas autotrophica (strain ATCC BAA-671 / DSM 16294 / JCM 11897 / OK10) OX=563040 GN=Saut_0591 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.32RR14 pKa = 11.84THH16 pKa = 6.0GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.32NGRR28 pKa = 11.84NIINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.99GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.21LTVV44 pKa = 3.1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.32RR14 pKa = 11.84THH16 pKa = 6.0GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.32NGRR28 pKa = 11.84NIINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.99GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.21LTVV44 pKa = 3.1
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
672986 |
30 |
1662 |
312.3 |
35.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.079 ± 0.053 | 0.858 ± 0.021 |
5.685 ± 0.033 | 6.933 ± 0.063 |
4.868 ± 0.043 | 5.729 ± 0.056 |
2.039 ± 0.021 | 8.356 ± 0.057 |
8.715 ± 0.06 | 9.749 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.693 ± 0.023 | 5.311 ± 0.047 |
2.971 ± 0.034 | 3.195 ± 0.03 |
3.265 ± 0.038 | 6.261 ± 0.043 |
5.121 ± 0.046 | 6.357 ± 0.041 |
0.731 ± 0.018 | 4.084 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
