
Klebsiella phage JD001
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Jedunavirus; Klebsiella virus JD001
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
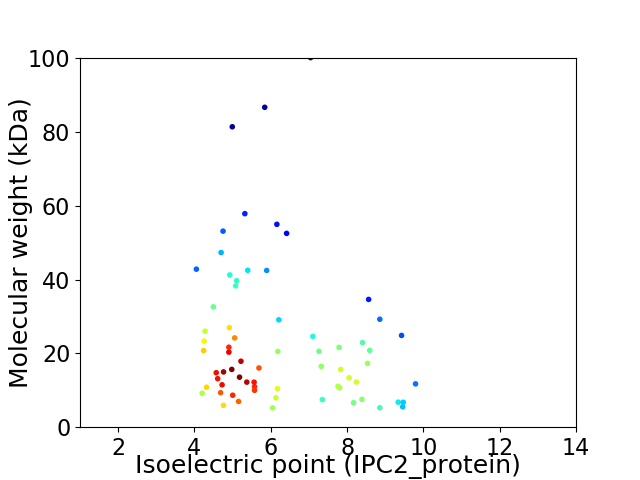
Virtual 2D-PAGE plot for 68 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0ARS9|L0ARS9_9CAUD Uncharacterized protein OS=Klebsiella phage JD001 OX=1236000 PE=4 SV=1
MM1 pKa = 7.92AEE3 pKa = 4.19LTSTGYY9 pKa = 9.8SVKK12 pKa = 10.61SQNDD16 pKa = 3.0WFDD19 pKa = 3.59EE20 pKa = 4.25EE21 pKa = 4.28KK22 pKa = 10.67QLYY25 pKa = 10.35LDD27 pKa = 5.34IDD29 pKa = 4.75SNWNLDD35 pKa = 3.15PSTPDD40 pKa = 3.22GLKK43 pKa = 9.64IAHH46 pKa = 6.95DD47 pKa = 3.98AEE49 pKa = 4.46IFSALDD55 pKa = 3.24EE56 pKa = 4.48VLQQAYY62 pKa = 10.17NSKK65 pKa = 10.83DD66 pKa = 3.36PNKK69 pKa = 10.71ASGYY73 pKa = 10.2DD74 pKa = 3.51LDD76 pKa = 4.94VICALTGTVRR86 pKa = 11.84SEE88 pKa = 4.03GTASTVTGFVLTGTPGTQVPAGTRR112 pKa = 11.84FEE114 pKa = 4.52SSVTGYY120 pKa = 10.93RR121 pKa = 11.84FTLDD125 pKa = 3.18QTWTLDD131 pKa = 3.37SSGTATVDD139 pKa = 3.23ITCTTVGEE147 pKa = 4.34IEE149 pKa = 4.58ADD151 pKa = 3.23ANTITTIVDD160 pKa = 3.73TVAGLVSVNNPTPATPGTSAEE181 pKa = 4.23SDD183 pKa = 3.05GSLRR187 pKa = 11.84LKK189 pKa = 10.49RR190 pKa = 11.84ATAVGLPGSNQVDD203 pKa = 3.73SMLGQLFNVDD213 pKa = 2.55GVRR216 pKa = 11.84RR217 pKa = 11.84VRR219 pKa = 11.84VYY221 pKa = 11.09EE222 pKa = 4.02NDD224 pKa = 3.25EE225 pKa = 4.03ATTDD229 pKa = 3.55SNGQPGHH236 pKa = 6.53SIAPIVDD243 pKa = 3.38GGTDD247 pKa = 3.87DD248 pKa = 3.93NVAMAIYY255 pKa = 9.96LKK257 pKa = 10.46KK258 pKa = 10.73NPGVTLYY265 pKa = 10.7QAGTPVTVTVTSPTYY280 pKa = 8.95PTMTKK285 pKa = 9.97DD286 pKa = 3.14IKK288 pKa = 10.47FSRR291 pKa = 11.84PVYY294 pKa = 9.2VDD296 pKa = 2.87MVVVIEE302 pKa = 4.51IKK304 pKa = 10.65DD305 pKa = 3.9DD306 pKa = 3.66GTLPSQATLEE316 pKa = 4.24PLIQDD321 pKa = 4.96AIMEE325 pKa = 4.3YY326 pKa = 10.66AAGGLIPTEE335 pKa = 4.23YY336 pKa = 10.73GFKK339 pKa = 10.12PDD341 pKa = 3.92GFDD344 pKa = 2.96IGEE347 pKa = 4.3TVPYY351 pKa = 10.09SSLYY355 pKa = 9.26TPINKK360 pKa = 9.51VIGSYY365 pKa = 10.9GNSYY369 pKa = 9.48VNSMTLNGGTANVTIDD385 pKa = 3.64FNEE388 pKa = 4.08LSRR391 pKa = 11.84WTTSNITVTIVV402 pKa = 2.6
MM1 pKa = 7.92AEE3 pKa = 4.19LTSTGYY9 pKa = 9.8SVKK12 pKa = 10.61SQNDD16 pKa = 3.0WFDD19 pKa = 3.59EE20 pKa = 4.25EE21 pKa = 4.28KK22 pKa = 10.67QLYY25 pKa = 10.35LDD27 pKa = 5.34IDD29 pKa = 4.75SNWNLDD35 pKa = 3.15PSTPDD40 pKa = 3.22GLKK43 pKa = 9.64IAHH46 pKa = 6.95DD47 pKa = 3.98AEE49 pKa = 4.46IFSALDD55 pKa = 3.24EE56 pKa = 4.48VLQQAYY62 pKa = 10.17NSKK65 pKa = 10.83DD66 pKa = 3.36PNKK69 pKa = 10.71ASGYY73 pKa = 10.2DD74 pKa = 3.51LDD76 pKa = 4.94VICALTGTVRR86 pKa = 11.84SEE88 pKa = 4.03GTASTVTGFVLTGTPGTQVPAGTRR112 pKa = 11.84FEE114 pKa = 4.52SSVTGYY120 pKa = 10.93RR121 pKa = 11.84FTLDD125 pKa = 3.18QTWTLDD131 pKa = 3.37SSGTATVDD139 pKa = 3.23ITCTTVGEE147 pKa = 4.34IEE149 pKa = 4.58ADD151 pKa = 3.23ANTITTIVDD160 pKa = 3.73TVAGLVSVNNPTPATPGTSAEE181 pKa = 4.23SDD183 pKa = 3.05GSLRR187 pKa = 11.84LKK189 pKa = 10.49RR190 pKa = 11.84ATAVGLPGSNQVDD203 pKa = 3.73SMLGQLFNVDD213 pKa = 2.55GVRR216 pKa = 11.84RR217 pKa = 11.84VRR219 pKa = 11.84VYY221 pKa = 11.09EE222 pKa = 4.02NDD224 pKa = 3.25EE225 pKa = 4.03ATTDD229 pKa = 3.55SNGQPGHH236 pKa = 6.53SIAPIVDD243 pKa = 3.38GGTDD247 pKa = 3.87DD248 pKa = 3.93NVAMAIYY255 pKa = 9.96LKK257 pKa = 10.46KK258 pKa = 10.73NPGVTLYY265 pKa = 10.7QAGTPVTVTVTSPTYY280 pKa = 8.95PTMTKK285 pKa = 9.97DD286 pKa = 3.14IKK288 pKa = 10.47FSRR291 pKa = 11.84PVYY294 pKa = 9.2VDD296 pKa = 2.87MVVVIEE302 pKa = 4.51IKK304 pKa = 10.65DD305 pKa = 3.9DD306 pKa = 3.66GTLPSQATLEE316 pKa = 4.24PLIQDD321 pKa = 4.96AIMEE325 pKa = 4.3YY326 pKa = 10.66AAGGLIPTEE335 pKa = 4.23YY336 pKa = 10.73GFKK339 pKa = 10.12PDD341 pKa = 3.92GFDD344 pKa = 2.96IGEE347 pKa = 4.3TVPYY351 pKa = 10.09SSLYY355 pKa = 9.26TPINKK360 pKa = 9.51VIGSYY365 pKa = 10.9GNSYY369 pKa = 9.48VNSMTLNGGTANVTIDD385 pKa = 3.64FNEE388 pKa = 4.08LSRR391 pKa = 11.84WTTSNITVTIVV402 pKa = 2.6
Molecular weight: 42.84 kDa
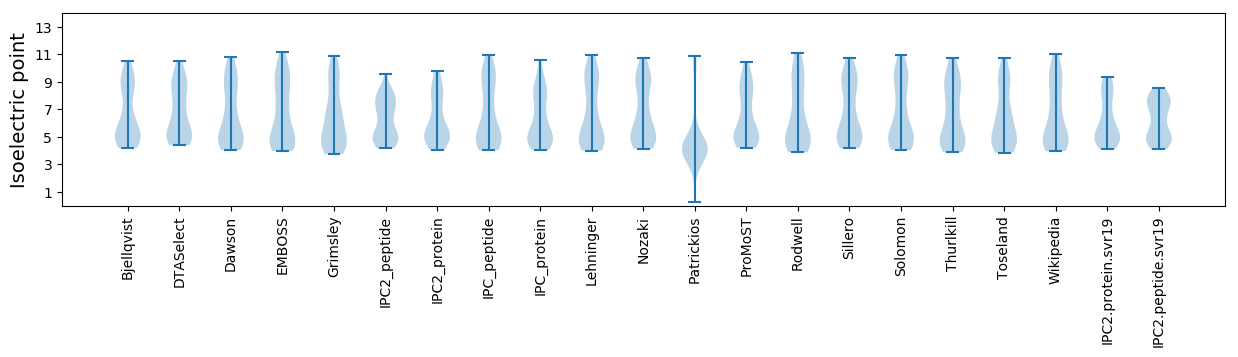
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0AQN9|L0AQN9_9CAUD Uncharacterized protein OS=Klebsiella phage JD001 OX=1236000 PE=4 SV=1
MM1 pKa = 7.49GSLVEE6 pKa = 5.26DD7 pKa = 4.22ILRR10 pKa = 11.84HH11 pKa = 5.25VPTKK15 pKa = 8.54QWRR18 pKa = 11.84KK19 pKa = 8.12KK20 pKa = 9.44CDD22 pKa = 3.71RR23 pKa = 11.84IASRR27 pKa = 11.84EE28 pKa = 3.89LHH30 pKa = 6.12LARR33 pKa = 11.84VPGIHH38 pKa = 6.4HH39 pKa = 7.56RR40 pKa = 11.84YY41 pKa = 8.84GKK43 pKa = 9.13WEE45 pKa = 3.77AVVFYY50 pKa = 11.0RR51 pKa = 11.84NVTLTIGCFNTPHH64 pKa = 7.12RR65 pKa = 11.84ALLARR70 pKa = 11.84KK71 pKa = 8.75LWYY74 pKa = 9.31FWRR77 pKa = 11.84EE78 pKa = 3.56RR79 pKa = 11.84GFTPQEE85 pKa = 3.75IPKK88 pKa = 9.88GPKK91 pKa = 8.95RR92 pKa = 11.84EE93 pKa = 4.08PYY95 pKa = 10.14SRR97 pKa = 11.84SS98 pKa = 3.19
MM1 pKa = 7.49GSLVEE6 pKa = 5.26DD7 pKa = 4.22ILRR10 pKa = 11.84HH11 pKa = 5.25VPTKK15 pKa = 8.54QWRR18 pKa = 11.84KK19 pKa = 8.12KK20 pKa = 9.44CDD22 pKa = 3.71RR23 pKa = 11.84IASRR27 pKa = 11.84EE28 pKa = 3.89LHH30 pKa = 6.12LARR33 pKa = 11.84VPGIHH38 pKa = 6.4HH39 pKa = 7.56RR40 pKa = 11.84YY41 pKa = 8.84GKK43 pKa = 9.13WEE45 pKa = 3.77AVVFYY50 pKa = 11.0RR51 pKa = 11.84NVTLTIGCFNTPHH64 pKa = 7.12RR65 pKa = 11.84ALLARR70 pKa = 11.84KK71 pKa = 8.75LWYY74 pKa = 9.31FWRR77 pKa = 11.84EE78 pKa = 3.56RR79 pKa = 11.84GFTPQEE85 pKa = 3.75IPKK88 pKa = 9.88GPKK91 pKa = 8.95RR92 pKa = 11.84EE93 pKa = 4.08PYY95 pKa = 10.14SRR97 pKa = 11.84SS98 pKa = 3.19
Molecular weight: 11.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14611 |
43 |
898 |
214.9 |
23.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.685 ± 0.471 | 1.383 ± 0.154 |
6.379 ± 0.199 | 5.496 ± 0.287 |
3.532 ± 0.192 | 7.672 ± 0.453 |
1.848 ± 0.165 | 5.893 ± 0.198 |
5.489 ± 0.342 | 7.535 ± 0.231 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.614 ± 0.14 | 4.88 ± 0.28 |
3.997 ± 0.249 | 3.887 ± 0.264 |
5.065 ± 0.212 | 6.146 ± 0.267 |
7.002 ± 0.359 | 7.262 ± 0.266 |
1.554 ± 0.106 | 3.682 ± 0.186 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
