
DPANN group archaeon LC1Nh
Taxonomy: cellular organisms; Archaea; DPANN group; Candidatus Nanohaloarchaeota; Candidatus Nanohalobia; Candidatus Nanohalobiales; Candidatus Nanohalobiaceae; Candidatus Nanohalobium
Average proteome isoelectric point is 5.13
Get precalculated fractions of proteins
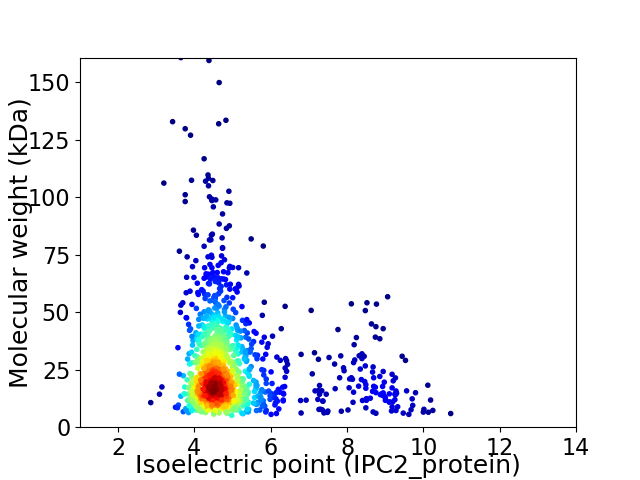
Virtual 2D-PAGE plot for 1162 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q0UFP3|A0A5Q0UFP3_9ARCH Uncharacterized protein OS=DPANN group archaeon LC1Nh OX=2565781 GN=LC1Nh_0518 PE=4 SV=1
MM1 pKa = 7.16KK2 pKa = 10.23RR3 pKa = 11.84KK4 pKa = 9.35IGSKK8 pKa = 9.64IGLLIVFAAALTSVAAAQATINYY31 pKa = 8.28DD32 pKa = 3.65YY33 pKa = 11.41NVDD36 pKa = 3.84DD37 pKa = 4.51VEE39 pKa = 5.26VMAFNCLNSTDD50 pKa = 4.06CSNVGDD56 pKa = 3.97FGGNILDD63 pKa = 5.05SGTTSNGEE71 pKa = 3.59ITIDD75 pKa = 3.79YY76 pKa = 8.9PAEE79 pKa = 4.22LQDD82 pKa = 3.41HH83 pKa = 6.99GYY85 pKa = 10.81AVYY88 pKa = 9.84HH89 pKa = 5.9VSPGYY94 pKa = 10.9VPVEE98 pKa = 4.65AYY100 pKa = 10.52ADD102 pKa = 3.87WSCGGSGSCGGNTTDD117 pKa = 3.78VQFEE121 pKa = 4.68KK122 pKa = 10.34ISQCSATVDD131 pKa = 3.73SFSVTNDD138 pKa = 2.66VYY140 pKa = 11.45ANEE143 pKa = 4.47PLTVDD148 pKa = 3.49VSATAEE154 pKa = 4.35STVSSAFEE162 pKa = 3.54EE163 pKa = 4.69AGPVDD168 pKa = 4.7HH169 pKa = 7.17VPEE172 pKa = 4.78AYY174 pKa = 9.5KK175 pKa = 10.59DD176 pKa = 3.47QYY178 pKa = 11.75YY179 pKa = 10.63SANTNISLEE188 pKa = 4.26VTDD191 pKa = 3.81TDD193 pKa = 3.97GNPVHH198 pKa = 6.57TAEE201 pKa = 4.11QQKK204 pKa = 10.23NIYY207 pKa = 9.09MNEE210 pKa = 3.81TDD212 pKa = 3.3QVEE215 pKa = 4.65FGWTPSKK222 pKa = 11.01SEE224 pKa = 4.03NYY226 pKa = 8.26TVQASTEE233 pKa = 4.27VIDD236 pKa = 4.44PQCSSTNILSTDD248 pKa = 3.4KK249 pKa = 11.24EE250 pKa = 4.63PITVHH255 pKa = 5.9EE256 pKa = 4.41EE257 pKa = 3.93RR258 pKa = 11.84PQNEE262 pKa = 5.22YY263 pKa = 9.33YY264 pKa = 10.65TLINDD269 pKa = 5.35LDD271 pKa = 3.85IQDD274 pKa = 4.1NHH276 pKa = 6.78PKK278 pKa = 9.26ATQQQTFTYY287 pKa = 10.31SKK289 pKa = 9.9ISNYY293 pKa = 10.36ADD295 pKa = 3.13NDD297 pKa = 3.55YY298 pKa = 11.51DD299 pKa = 3.66KK300 pKa = 10.76TAVKK304 pKa = 9.97TDD306 pKa = 2.98VNYY309 pKa = 9.54TVRR312 pKa = 11.84DD313 pKa = 3.37SDD315 pKa = 3.74GQEE318 pKa = 3.96VNFSTEE324 pKa = 3.69QLPANQNEE332 pKa = 3.82QDD334 pKa = 3.56AEE336 pKa = 4.32SFSFSWTPKK345 pKa = 9.23SAGTYY350 pKa = 6.33TVEE353 pKa = 4.12VAGEE357 pKa = 4.23SVQTEE362 pKa = 4.68GEE364 pKa = 4.2NPPTSPDD371 pKa = 3.43TANLEE376 pKa = 3.62IDD378 pKa = 3.75VAEE381 pKa = 4.12EE382 pKa = 4.09PKK384 pKa = 10.35WKK386 pKa = 10.85VEE388 pKa = 4.03FNVVDD393 pKa = 3.83TNGDD397 pKa = 3.59PVNNAEE403 pKa = 4.29VKK405 pKa = 10.74INDD408 pKa = 3.85RR409 pKa = 11.84TNQTDD414 pKa = 3.58SNGNTEE420 pKa = 3.83ITKK423 pKa = 8.78LTKK426 pKa = 10.25GSYY429 pKa = 8.61TFNVSKK435 pKa = 9.66TGYY438 pKa = 5.27TTSSGSLTIEE448 pKa = 4.26STTSDD453 pKa = 4.03PLQKK457 pKa = 10.58SVVIQNNNTAPKK469 pKa = 10.69VLDD472 pKa = 3.64IPNKK476 pKa = 10.26KK477 pKa = 9.36IVEE480 pKa = 4.33GDD482 pKa = 3.52TNNDD486 pKa = 3.0INLDD490 pKa = 3.93NYY492 pKa = 10.7VSDD495 pKa = 4.14QQDD498 pKa = 3.49LASEE502 pKa = 4.41ISWTSNGAEE511 pKa = 4.32DD512 pKa = 4.81NQNLDD517 pKa = 3.45VNIDD521 pKa = 3.79PDD523 pKa = 3.75THH525 pKa = 6.76DD526 pKa = 3.8VNITSTSGWTGTEE539 pKa = 4.23SITFTATDD547 pKa = 3.43TAGATDD553 pKa = 4.22SDD555 pKa = 4.51SMTVTVTEE563 pKa = 4.63KK564 pKa = 9.73NTAPTVSEE572 pKa = 4.32IPDD575 pKa = 3.35VTLDD579 pKa = 3.76EE580 pKa = 4.86DD581 pKa = 3.92TTEE584 pKa = 3.76YY585 pKa = 11.12DD586 pKa = 3.54YY587 pKa = 12.17VNLVNFVSDD596 pKa = 5.07AEE598 pKa = 4.37STYY601 pKa = 11.56NNLDD605 pKa = 3.06VSIDD609 pKa = 3.68VSSDD613 pKa = 2.88KK614 pKa = 11.14CGVSIDD620 pKa = 3.92EE621 pKa = 4.44NEE623 pKa = 3.81IDD625 pKa = 4.27IAPNTNFHH633 pKa = 5.71GTCTVTVQATDD644 pKa = 3.8PYY646 pKa = 11.46GATTSNNFEE655 pKa = 3.97ITVNPVNDD663 pKa = 4.07PPTITSNPQTTVDD676 pKa = 4.84AYY678 pKa = 8.6TQYY681 pKa = 11.28YY682 pKa = 8.39YY683 pKa = 10.67QLSVSDD689 pKa = 4.71PDD691 pKa = 3.32NGAASFTYY699 pKa = 9.79TKK701 pKa = 10.32EE702 pKa = 3.7QGPANMTINQNGEE715 pKa = 4.39VTWTPTNNNADD726 pKa = 3.88NSPHH730 pKa = 5.95QVTLNVSDD738 pKa = 4.03GTNSTTQSYY747 pKa = 11.22SLTVNKK753 pKa = 10.13APKK756 pKa = 9.54NQIPHH761 pKa = 6.82PEE763 pKa = 3.52ITVNKK768 pKa = 8.71TKK770 pKa = 10.69GKK772 pKa = 10.79APLTVNFNASNSADD786 pKa = 3.62PDD788 pKa = 3.96GSITQYY794 pKa = 10.35NWSFGEE800 pKa = 4.13GGKK803 pKa = 10.78ANGIEE808 pKa = 4.14TNYY811 pKa = 10.62TYY813 pKa = 8.96TQPGNYY819 pKa = 7.65TATLTLTDD827 pKa = 4.9DD828 pKa = 4.08NGSTATEE835 pKa = 4.04SVRR838 pKa = 11.84IEE840 pKa = 3.94VTGNNAPSFTSTSFKK855 pKa = 8.51TTATVNQQYY864 pKa = 8.89TYY866 pKa = 10.69DD867 pKa = 3.82AQATDD872 pKa = 3.8PDD874 pKa = 4.83GDD876 pKa = 3.81TLEE879 pKa = 4.61YY880 pKa = 10.84SLNTAPQSMSINNNTGKK897 pKa = 8.47ITWTPSNTGEE907 pKa = 4.24YY908 pKa = 9.65KK909 pKa = 10.63VEE911 pKa = 3.91IQVSDD916 pKa = 3.72GTDD919 pKa = 3.1TAIQSYY925 pKa = 10.47NITVSSNSDD934 pKa = 3.08NDD936 pKa = 3.9GGGDD940 pKa = 3.85DD941 pKa = 4.51EE942 pKa = 6.23DD943 pKa = 4.78SGGSSSGGGGGGGGGGFGGSFTTTDD968 pKa = 3.42EE969 pKa = 4.32EE970 pKa = 4.45EE971 pKa = 4.22EE972 pKa = 4.0EE973 pKa = 4.44SIEE976 pKa = 4.52EE977 pKa = 4.52RR978 pKa = 11.84IQTGDD983 pKa = 3.59NEE985 pKa = 4.18VDD987 pKa = 3.65LDD989 pKa = 4.09IEE991 pKa = 4.38PGIDD995 pKa = 3.38DD996 pKa = 4.79SYY998 pKa = 11.95NLTFNVSEE1006 pKa = 4.5DD1007 pKa = 3.86TNITTSVTGNVSEE1020 pKa = 4.42IVSVPANFTAKK1031 pKa = 10.29KK1032 pKa = 10.51GEE1034 pKa = 4.27NNLPLQINGSVPGNYY1049 pKa = 8.89TGNLTLQWNNQTLNIPLNLNIEE1071 pKa = 4.35DD1072 pKa = 4.31TGASDD1077 pKa = 3.72YY1078 pKa = 11.56SVAILHH1084 pKa = 6.68KK1085 pKa = 9.13STADD1089 pKa = 3.23EE1090 pKa = 4.68RR1091 pKa = 11.84ISYY1094 pKa = 9.44DD1095 pKa = 3.17VKK1097 pKa = 10.9LWDD1100 pKa = 3.88EE1101 pKa = 4.48EE1102 pKa = 4.59KK1103 pKa = 10.54PDD1105 pKa = 3.51TSNQVSITTSVIDD1118 pKa = 3.64SEE1120 pKa = 4.33GDD1122 pKa = 3.08EE1123 pKa = 4.42VYY1125 pKa = 10.91RR1126 pKa = 11.84NTEE1129 pKa = 3.54TEE1131 pKa = 3.86RR1132 pKa = 11.84IYY1134 pKa = 11.3DD1135 pKa = 3.57VLVRR1139 pKa = 11.84NYY1141 pKa = 7.73RR1142 pKa = 11.84TSILPEE1148 pKa = 3.85GEE1150 pKa = 4.25YY1151 pKa = 10.33TVTATVTDD1159 pKa = 4.21DD1160 pKa = 3.88GEE1162 pKa = 4.4EE1163 pKa = 4.2FTGSRR1168 pKa = 11.84TIQLGPSEE1176 pKa = 4.7TITGQFTSSNSLGSLFQDD1194 pKa = 4.07IIDD1197 pKa = 4.58SISNLFF1203 pKa = 3.51
MM1 pKa = 7.16KK2 pKa = 10.23RR3 pKa = 11.84KK4 pKa = 9.35IGSKK8 pKa = 9.64IGLLIVFAAALTSVAAAQATINYY31 pKa = 8.28DD32 pKa = 3.65YY33 pKa = 11.41NVDD36 pKa = 3.84DD37 pKa = 4.51VEE39 pKa = 5.26VMAFNCLNSTDD50 pKa = 4.06CSNVGDD56 pKa = 3.97FGGNILDD63 pKa = 5.05SGTTSNGEE71 pKa = 3.59ITIDD75 pKa = 3.79YY76 pKa = 8.9PAEE79 pKa = 4.22LQDD82 pKa = 3.41HH83 pKa = 6.99GYY85 pKa = 10.81AVYY88 pKa = 9.84HH89 pKa = 5.9VSPGYY94 pKa = 10.9VPVEE98 pKa = 4.65AYY100 pKa = 10.52ADD102 pKa = 3.87WSCGGSGSCGGNTTDD117 pKa = 3.78VQFEE121 pKa = 4.68KK122 pKa = 10.34ISQCSATVDD131 pKa = 3.73SFSVTNDD138 pKa = 2.66VYY140 pKa = 11.45ANEE143 pKa = 4.47PLTVDD148 pKa = 3.49VSATAEE154 pKa = 4.35STVSSAFEE162 pKa = 3.54EE163 pKa = 4.69AGPVDD168 pKa = 4.7HH169 pKa = 7.17VPEE172 pKa = 4.78AYY174 pKa = 9.5KK175 pKa = 10.59DD176 pKa = 3.47QYY178 pKa = 11.75YY179 pKa = 10.63SANTNISLEE188 pKa = 4.26VTDD191 pKa = 3.81TDD193 pKa = 3.97GNPVHH198 pKa = 6.57TAEE201 pKa = 4.11QQKK204 pKa = 10.23NIYY207 pKa = 9.09MNEE210 pKa = 3.81TDD212 pKa = 3.3QVEE215 pKa = 4.65FGWTPSKK222 pKa = 11.01SEE224 pKa = 4.03NYY226 pKa = 8.26TVQASTEE233 pKa = 4.27VIDD236 pKa = 4.44PQCSSTNILSTDD248 pKa = 3.4KK249 pKa = 11.24EE250 pKa = 4.63PITVHH255 pKa = 5.9EE256 pKa = 4.41EE257 pKa = 3.93RR258 pKa = 11.84PQNEE262 pKa = 5.22YY263 pKa = 9.33YY264 pKa = 10.65TLINDD269 pKa = 5.35LDD271 pKa = 3.85IQDD274 pKa = 4.1NHH276 pKa = 6.78PKK278 pKa = 9.26ATQQQTFTYY287 pKa = 10.31SKK289 pKa = 9.9ISNYY293 pKa = 10.36ADD295 pKa = 3.13NDD297 pKa = 3.55YY298 pKa = 11.51DD299 pKa = 3.66KK300 pKa = 10.76TAVKK304 pKa = 9.97TDD306 pKa = 2.98VNYY309 pKa = 9.54TVRR312 pKa = 11.84DD313 pKa = 3.37SDD315 pKa = 3.74GQEE318 pKa = 3.96VNFSTEE324 pKa = 3.69QLPANQNEE332 pKa = 3.82QDD334 pKa = 3.56AEE336 pKa = 4.32SFSFSWTPKK345 pKa = 9.23SAGTYY350 pKa = 6.33TVEE353 pKa = 4.12VAGEE357 pKa = 4.23SVQTEE362 pKa = 4.68GEE364 pKa = 4.2NPPTSPDD371 pKa = 3.43TANLEE376 pKa = 3.62IDD378 pKa = 3.75VAEE381 pKa = 4.12EE382 pKa = 4.09PKK384 pKa = 10.35WKK386 pKa = 10.85VEE388 pKa = 4.03FNVVDD393 pKa = 3.83TNGDD397 pKa = 3.59PVNNAEE403 pKa = 4.29VKK405 pKa = 10.74INDD408 pKa = 3.85RR409 pKa = 11.84TNQTDD414 pKa = 3.58SNGNTEE420 pKa = 3.83ITKK423 pKa = 8.78LTKK426 pKa = 10.25GSYY429 pKa = 8.61TFNVSKK435 pKa = 9.66TGYY438 pKa = 5.27TTSSGSLTIEE448 pKa = 4.26STTSDD453 pKa = 4.03PLQKK457 pKa = 10.58SVVIQNNNTAPKK469 pKa = 10.69VLDD472 pKa = 3.64IPNKK476 pKa = 10.26KK477 pKa = 9.36IVEE480 pKa = 4.33GDD482 pKa = 3.52TNNDD486 pKa = 3.0INLDD490 pKa = 3.93NYY492 pKa = 10.7VSDD495 pKa = 4.14QQDD498 pKa = 3.49LASEE502 pKa = 4.41ISWTSNGAEE511 pKa = 4.32DD512 pKa = 4.81NQNLDD517 pKa = 3.45VNIDD521 pKa = 3.79PDD523 pKa = 3.75THH525 pKa = 6.76DD526 pKa = 3.8VNITSTSGWTGTEE539 pKa = 4.23SITFTATDD547 pKa = 3.43TAGATDD553 pKa = 4.22SDD555 pKa = 4.51SMTVTVTEE563 pKa = 4.63KK564 pKa = 9.73NTAPTVSEE572 pKa = 4.32IPDD575 pKa = 3.35VTLDD579 pKa = 3.76EE580 pKa = 4.86DD581 pKa = 3.92TTEE584 pKa = 3.76YY585 pKa = 11.12DD586 pKa = 3.54YY587 pKa = 12.17VNLVNFVSDD596 pKa = 5.07AEE598 pKa = 4.37STYY601 pKa = 11.56NNLDD605 pKa = 3.06VSIDD609 pKa = 3.68VSSDD613 pKa = 2.88KK614 pKa = 11.14CGVSIDD620 pKa = 3.92EE621 pKa = 4.44NEE623 pKa = 3.81IDD625 pKa = 4.27IAPNTNFHH633 pKa = 5.71GTCTVTVQATDD644 pKa = 3.8PYY646 pKa = 11.46GATTSNNFEE655 pKa = 3.97ITVNPVNDD663 pKa = 4.07PPTITSNPQTTVDD676 pKa = 4.84AYY678 pKa = 8.6TQYY681 pKa = 11.28YY682 pKa = 8.39YY683 pKa = 10.67QLSVSDD689 pKa = 4.71PDD691 pKa = 3.32NGAASFTYY699 pKa = 9.79TKK701 pKa = 10.32EE702 pKa = 3.7QGPANMTINQNGEE715 pKa = 4.39VTWTPTNNNADD726 pKa = 3.88NSPHH730 pKa = 5.95QVTLNVSDD738 pKa = 4.03GTNSTTQSYY747 pKa = 11.22SLTVNKK753 pKa = 10.13APKK756 pKa = 9.54NQIPHH761 pKa = 6.82PEE763 pKa = 3.52ITVNKK768 pKa = 8.71TKK770 pKa = 10.69GKK772 pKa = 10.79APLTVNFNASNSADD786 pKa = 3.62PDD788 pKa = 3.96GSITQYY794 pKa = 10.35NWSFGEE800 pKa = 4.13GGKK803 pKa = 10.78ANGIEE808 pKa = 4.14TNYY811 pKa = 10.62TYY813 pKa = 8.96TQPGNYY819 pKa = 7.65TATLTLTDD827 pKa = 4.9DD828 pKa = 4.08NGSTATEE835 pKa = 4.04SVRR838 pKa = 11.84IEE840 pKa = 3.94VTGNNAPSFTSTSFKK855 pKa = 8.51TTATVNQQYY864 pKa = 8.89TYY866 pKa = 10.69DD867 pKa = 3.82AQATDD872 pKa = 3.8PDD874 pKa = 4.83GDD876 pKa = 3.81TLEE879 pKa = 4.61YY880 pKa = 10.84SLNTAPQSMSINNNTGKK897 pKa = 8.47ITWTPSNTGEE907 pKa = 4.24YY908 pKa = 9.65KK909 pKa = 10.63VEE911 pKa = 3.91IQVSDD916 pKa = 3.72GTDD919 pKa = 3.1TAIQSYY925 pKa = 10.47NITVSSNSDD934 pKa = 3.08NDD936 pKa = 3.9GGGDD940 pKa = 3.85DD941 pKa = 4.51EE942 pKa = 6.23DD943 pKa = 4.78SGGSSSGGGGGGGGGGFGGSFTTTDD968 pKa = 3.42EE969 pKa = 4.32EE970 pKa = 4.45EE971 pKa = 4.22EE972 pKa = 4.0EE973 pKa = 4.44SIEE976 pKa = 4.52EE977 pKa = 4.52RR978 pKa = 11.84IQTGDD983 pKa = 3.59NEE985 pKa = 4.18VDD987 pKa = 3.65LDD989 pKa = 4.09IEE991 pKa = 4.38PGIDD995 pKa = 3.38DD996 pKa = 4.79SYY998 pKa = 11.95NLTFNVSEE1006 pKa = 4.5DD1007 pKa = 3.86TNITTSVTGNVSEE1020 pKa = 4.42IVSVPANFTAKK1031 pKa = 10.29KK1032 pKa = 10.51GEE1034 pKa = 4.27NNLPLQINGSVPGNYY1049 pKa = 8.89TGNLTLQWNNQTLNIPLNLNIEE1071 pKa = 4.35DD1072 pKa = 4.31TGASDD1077 pKa = 3.72YY1078 pKa = 11.56SVAILHH1084 pKa = 6.68KK1085 pKa = 9.13STADD1089 pKa = 3.23EE1090 pKa = 4.68RR1091 pKa = 11.84ISYY1094 pKa = 9.44DD1095 pKa = 3.17VKK1097 pKa = 10.9LWDD1100 pKa = 3.88EE1101 pKa = 4.48EE1102 pKa = 4.59KK1103 pKa = 10.54PDD1105 pKa = 3.51TSNQVSITTSVIDD1118 pKa = 3.64SEE1120 pKa = 4.33GDD1122 pKa = 3.08EE1123 pKa = 4.42VYY1125 pKa = 10.91RR1126 pKa = 11.84NTEE1129 pKa = 3.54TEE1131 pKa = 3.86RR1132 pKa = 11.84IYY1134 pKa = 11.3DD1135 pKa = 3.57VLVRR1139 pKa = 11.84NYY1141 pKa = 7.73RR1142 pKa = 11.84TSILPEE1148 pKa = 3.85GEE1150 pKa = 4.25YY1151 pKa = 10.33TVTATVTDD1159 pKa = 4.21DD1160 pKa = 3.88GEE1162 pKa = 4.4EE1163 pKa = 4.2FTGSRR1168 pKa = 11.84TIQLGPSEE1176 pKa = 4.7TITGQFTSSNSLGSLFQDD1194 pKa = 4.07IIDD1197 pKa = 4.58SISNLFF1203 pKa = 3.51
Molecular weight: 129.72 kDa
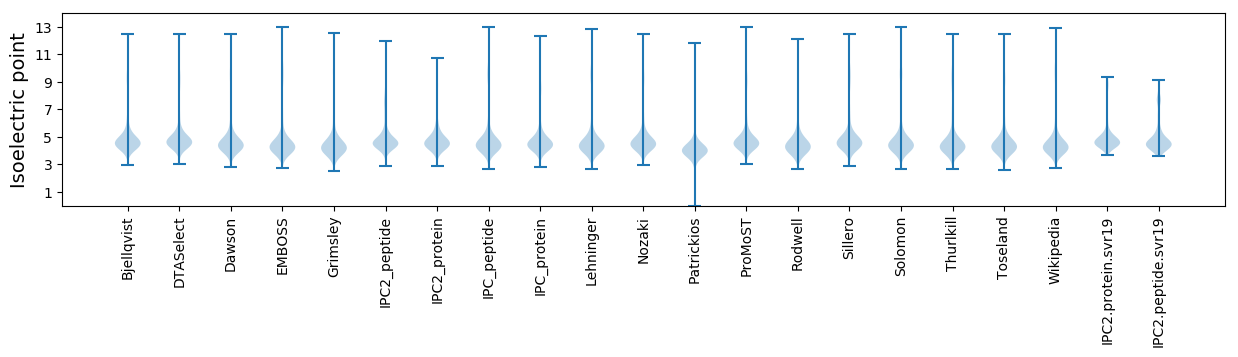
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q0UG78|A0A5Q0UG78_9ARCH ABC-2 type transport system ATP-binding protein OS=DPANN group archaeon LC1Nh OX=2565781 GN=LC1Nh_0707 PE=4 SV=1
MM1 pKa = 6.48NTKK4 pKa = 10.06NGRR7 pKa = 11.84KK8 pKa = 7.59RR9 pKa = 11.84TPLRR13 pKa = 11.84SRR15 pKa = 11.84VRR17 pKa = 11.84STALRR22 pKa = 11.84TRR24 pKa = 11.84TRR26 pKa = 11.84SNTSSRR32 pKa = 11.84RR33 pKa = 11.84HH34 pKa = 5.91RR35 pKa = 11.84SRR37 pKa = 11.84SSNSSLTKK45 pKa = 10.14IEE47 pKa = 4.09VLQDD51 pKa = 3.13
MM1 pKa = 6.48NTKK4 pKa = 10.06NGRR7 pKa = 11.84KK8 pKa = 7.59RR9 pKa = 11.84TPLRR13 pKa = 11.84SRR15 pKa = 11.84VRR17 pKa = 11.84STALRR22 pKa = 11.84TRR24 pKa = 11.84TRR26 pKa = 11.84SNTSSRR32 pKa = 11.84RR33 pKa = 11.84HH34 pKa = 5.91RR35 pKa = 11.84SRR37 pKa = 11.84SSNSSLTKK45 pKa = 10.14IEE47 pKa = 4.09VLQDD51 pKa = 3.13
Molecular weight: 5.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
293371 |
45 |
1499 |
252.5 |
28.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.045 ± 0.065 | 0.648 ± 0.026 |
7.212 ± 0.094 | 11.403 ± 0.155 |
3.968 ± 0.061 | 7.006 ± 0.079 |
1.674 ± 0.031 | 6.33 ± 0.064 |
6.339 ± 0.095 | 8.179 ± 0.093 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.312 ± 0.04 | 4.652 ± 0.072 |
3.258 ± 0.039 | 3.637 ± 0.055 |
4.38 ± 0.05 | 6.912 ± 0.092 |
5.307 ± 0.083 | 6.723 ± 0.064 |
0.879 ± 0.025 | 3.136 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
