
Neodiprion abietis NPV
Taxonomy: Viruses; Naldaviricetes; Lefavirales; Baculoviridae; Gammabaculovirus; unclassified Gammabaculovirus
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
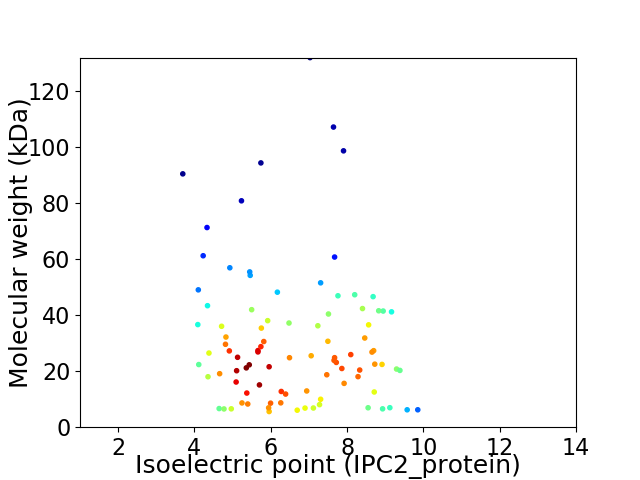
Virtual 2D-PAGE plot for 93 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q0ZNZ4|Q0ZNZ4_9CBAC Uncharacterized protein OS=Neodiprion abietis NPV OX=204507 PE=4 SV=1
MM1 pKa = 7.15FLIVFVFLCILLIFFTHH18 pKa = 6.78NIFVEE23 pKa = 4.5TTDD26 pKa = 5.28DD27 pKa = 3.41VTKK30 pKa = 10.85YY31 pKa = 10.56KK32 pKa = 10.47QIHH35 pKa = 5.9NLLLEE40 pKa = 5.03AYY42 pKa = 8.66TSDD45 pKa = 3.44LVSFDD50 pKa = 3.38TVSFISRR57 pKa = 11.84VRR59 pKa = 11.84WPVYY63 pKa = 9.95EE64 pKa = 3.67IVTFEE69 pKa = 4.26TEE71 pKa = 4.36TYY73 pKa = 10.93TMTNNMWYY81 pKa = 10.22SGLEE85 pKa = 3.72KK86 pKa = 10.51RR87 pKa = 11.84FNFYY91 pKa = 10.83NQTFEE96 pKa = 5.61DD97 pKa = 3.56VDD99 pKa = 4.26DD100 pKa = 4.07NAEE103 pKa = 3.96RR104 pKa = 11.84VTTVDD109 pKa = 3.08STSFSLHH116 pKa = 6.72VDD118 pKa = 3.78DD119 pKa = 5.42GWVNVDD125 pKa = 4.36CPANMIFDD133 pKa = 5.21GYY135 pKa = 10.73QCVRR139 pKa = 11.84QNICEE144 pKa = 4.03NQTAGTILPVNEE156 pKa = 4.32ALIDD160 pKa = 3.4VAFYY164 pKa = 10.92QKK166 pKa = 10.9YY167 pKa = 9.77NRR169 pKa = 11.84STTTDD174 pKa = 3.78TIYY177 pKa = 10.88HH178 pKa = 5.49PTLYY182 pKa = 9.83IVCNGDD188 pKa = 2.98DD189 pKa = 3.36TYY191 pKa = 11.36EE192 pKa = 4.78IKK194 pKa = 10.8EE195 pKa = 4.22CDD197 pKa = 3.35NNQLFDD203 pKa = 4.76SEE205 pKa = 4.53MLQCVDD211 pKa = 4.57KK212 pKa = 11.03DD213 pKa = 3.45ACADD217 pKa = 3.49VADD220 pKa = 4.28YY221 pKa = 10.35FVLSDD226 pKa = 4.96RR227 pKa = 11.84PDD229 pKa = 3.29TLADD233 pKa = 4.4DD234 pKa = 4.76EE235 pKa = 5.75YY236 pKa = 11.89LMCLNNEE243 pKa = 4.34TTVQTCEE250 pKa = 3.53EE251 pKa = 3.97NMIFDD256 pKa = 4.43TTTLSCVEE264 pKa = 4.05VDD266 pKa = 3.67LCQANGVGYY275 pKa = 8.32TYY277 pKa = 9.62ITDD280 pKa = 4.1DD281 pKa = 3.74LAANQYY287 pKa = 8.4YY288 pKa = 9.75RR289 pKa = 11.84CTSVFDD295 pKa = 4.25KK296 pKa = 10.98EE297 pKa = 4.8IVSCPIRR304 pKa = 11.84VEE306 pKa = 3.72TDD308 pKa = 3.01GQYY311 pKa = 11.11SCAGDD316 pKa = 3.92TQCMSLEE323 pKa = 4.27NGSGSIVTMTEE334 pKa = 3.6NTHH337 pKa = 6.25FKK339 pKa = 9.89YY340 pKa = 10.75ASEE343 pKa = 4.14AVICTEE349 pKa = 4.3YY350 pKa = 10.72EE351 pKa = 3.76ITDD354 pKa = 4.14SIVCDD359 pKa = 3.72SEE361 pKa = 4.64EE362 pKa = 4.28VEE364 pKa = 4.75LSVEE368 pKa = 4.22PFTLDD373 pKa = 3.43LKK375 pKa = 10.97IPTTILDD382 pKa = 3.59RR383 pKa = 11.84TNNVCKK389 pKa = 10.06TFDD392 pKa = 3.49INDD395 pKa = 3.32VEE397 pKa = 5.22IITDD401 pKa = 3.37YY402 pKa = 11.59AGIVNSEE409 pKa = 3.3NDD411 pKa = 3.5YY412 pKa = 11.57NVDD415 pKa = 3.34FTTAVRR421 pKa = 11.84VNVQNLLSEE430 pKa = 4.39NLPVSTSMLFSSLDD444 pKa = 3.32YY445 pKa = 10.88AKK447 pKa = 10.67NYY449 pKa = 10.01GFIGFSPLTGNEE461 pKa = 3.57IDD463 pKa = 4.34CYY465 pKa = 11.34GDD467 pKa = 3.13ILFDD471 pKa = 3.61IFAGDD476 pKa = 3.85KK477 pKa = 11.29YY478 pKa = 11.08NICSDD483 pKa = 3.91NEE485 pKa = 4.04ISSSNAIDD493 pKa = 4.02DD494 pKa = 4.24NEE496 pKa = 4.13FFSTYY501 pKa = 8.04TKK503 pKa = 11.03NVGTTTLTSPCRR515 pKa = 11.84VKK517 pKa = 10.98SDD519 pKa = 3.15IVLEE523 pKa = 4.04DD524 pKa = 4.88DD525 pKa = 4.88IYY527 pKa = 10.99IQEE530 pKa = 4.14SDD532 pKa = 3.74NVSVSCLYY540 pKa = 11.18AMPLQEE546 pKa = 5.9IIVTDD551 pKa = 4.04LEE553 pKa = 4.13EE554 pKa = 4.18AAGVLIDD561 pKa = 5.45DD562 pKa = 4.45KK563 pKa = 11.29CSEE566 pKa = 4.45NGIDD570 pKa = 3.87LNNVAGAIVILNDD583 pKa = 3.2EE584 pKa = 4.31LACRR588 pKa = 11.84SVYY591 pKa = 10.89LEE593 pKa = 4.92DD594 pKa = 3.72YY595 pKa = 10.71NVIITASFTPTMKK608 pKa = 10.76FEE610 pKa = 4.59DD611 pKa = 3.96RR612 pKa = 11.84TWNVDD617 pKa = 3.19TQSYY621 pKa = 10.09DD622 pKa = 2.77GVMYY626 pKa = 10.53DD627 pKa = 3.92SVVHH631 pKa = 6.89KK632 pKa = 10.23ITDD635 pKa = 2.99GWMACPPSAYY645 pKa = 10.33DD646 pKa = 4.93KK647 pKa = 8.26TTTDD651 pKa = 3.98CNIDD655 pKa = 3.36NTKK658 pKa = 10.77LYY660 pKa = 9.11TVKK663 pKa = 10.61DD664 pKa = 3.16LHH666 pKa = 6.65FVQNYY671 pKa = 9.27PDD673 pKa = 3.61IDD675 pKa = 3.91YY676 pKa = 11.07EE677 pKa = 4.21PEE679 pKa = 3.81NEE681 pKa = 4.26VNVDD685 pKa = 3.61TEE687 pKa = 4.8DD688 pKa = 3.75VDD690 pKa = 3.98VSNPIADD697 pKa = 3.88DD698 pKa = 3.82VEE700 pKa = 5.29AINPIAEE707 pKa = 4.22DD708 pKa = 3.52VLRR711 pKa = 11.84QNDD714 pKa = 3.46NVTNNVQPALEE725 pKa = 4.23NEE727 pKa = 4.6LVNTATQMEE736 pKa = 4.66VDD738 pKa = 3.87QKK740 pKa = 11.24NPSINVQDD748 pKa = 3.92IDD750 pKa = 4.18SSSTLSDD757 pKa = 3.06EE758 pKa = 4.54SVKK761 pKa = 10.78NDD763 pKa = 3.41YY764 pKa = 9.11STLDD768 pKa = 3.28TNRR771 pKa = 11.84ILDD774 pKa = 4.06YY775 pKa = 10.77INRR778 pKa = 11.84HH779 pKa = 5.78HH780 pKa = 6.86EE781 pKa = 4.28SEE783 pKa = 4.36TSEE786 pKa = 4.28EE787 pKa = 4.46EE788 pKa = 4.14YY789 pKa = 10.48MDD791 pKa = 5.57DD792 pKa = 4.73GDD794 pKa = 4.04ILASLFEE801 pKa = 4.25
MM1 pKa = 7.15FLIVFVFLCILLIFFTHH18 pKa = 6.78NIFVEE23 pKa = 4.5TTDD26 pKa = 5.28DD27 pKa = 3.41VTKK30 pKa = 10.85YY31 pKa = 10.56KK32 pKa = 10.47QIHH35 pKa = 5.9NLLLEE40 pKa = 5.03AYY42 pKa = 8.66TSDD45 pKa = 3.44LVSFDD50 pKa = 3.38TVSFISRR57 pKa = 11.84VRR59 pKa = 11.84WPVYY63 pKa = 9.95EE64 pKa = 3.67IVTFEE69 pKa = 4.26TEE71 pKa = 4.36TYY73 pKa = 10.93TMTNNMWYY81 pKa = 10.22SGLEE85 pKa = 3.72KK86 pKa = 10.51RR87 pKa = 11.84FNFYY91 pKa = 10.83NQTFEE96 pKa = 5.61DD97 pKa = 3.56VDD99 pKa = 4.26DD100 pKa = 4.07NAEE103 pKa = 3.96RR104 pKa = 11.84VTTVDD109 pKa = 3.08STSFSLHH116 pKa = 6.72VDD118 pKa = 3.78DD119 pKa = 5.42GWVNVDD125 pKa = 4.36CPANMIFDD133 pKa = 5.21GYY135 pKa = 10.73QCVRR139 pKa = 11.84QNICEE144 pKa = 4.03NQTAGTILPVNEE156 pKa = 4.32ALIDD160 pKa = 3.4VAFYY164 pKa = 10.92QKK166 pKa = 10.9YY167 pKa = 9.77NRR169 pKa = 11.84STTTDD174 pKa = 3.78TIYY177 pKa = 10.88HH178 pKa = 5.49PTLYY182 pKa = 9.83IVCNGDD188 pKa = 2.98DD189 pKa = 3.36TYY191 pKa = 11.36EE192 pKa = 4.78IKK194 pKa = 10.8EE195 pKa = 4.22CDD197 pKa = 3.35NNQLFDD203 pKa = 4.76SEE205 pKa = 4.53MLQCVDD211 pKa = 4.57KK212 pKa = 11.03DD213 pKa = 3.45ACADD217 pKa = 3.49VADD220 pKa = 4.28YY221 pKa = 10.35FVLSDD226 pKa = 4.96RR227 pKa = 11.84PDD229 pKa = 3.29TLADD233 pKa = 4.4DD234 pKa = 4.76EE235 pKa = 5.75YY236 pKa = 11.89LMCLNNEE243 pKa = 4.34TTVQTCEE250 pKa = 3.53EE251 pKa = 3.97NMIFDD256 pKa = 4.43TTTLSCVEE264 pKa = 4.05VDD266 pKa = 3.67LCQANGVGYY275 pKa = 8.32TYY277 pKa = 9.62ITDD280 pKa = 4.1DD281 pKa = 3.74LAANQYY287 pKa = 8.4YY288 pKa = 9.75RR289 pKa = 11.84CTSVFDD295 pKa = 4.25KK296 pKa = 10.98EE297 pKa = 4.8IVSCPIRR304 pKa = 11.84VEE306 pKa = 3.72TDD308 pKa = 3.01GQYY311 pKa = 11.11SCAGDD316 pKa = 3.92TQCMSLEE323 pKa = 4.27NGSGSIVTMTEE334 pKa = 3.6NTHH337 pKa = 6.25FKK339 pKa = 9.89YY340 pKa = 10.75ASEE343 pKa = 4.14AVICTEE349 pKa = 4.3YY350 pKa = 10.72EE351 pKa = 3.76ITDD354 pKa = 4.14SIVCDD359 pKa = 3.72SEE361 pKa = 4.64EE362 pKa = 4.28VEE364 pKa = 4.75LSVEE368 pKa = 4.22PFTLDD373 pKa = 3.43LKK375 pKa = 10.97IPTTILDD382 pKa = 3.59RR383 pKa = 11.84TNNVCKK389 pKa = 10.06TFDD392 pKa = 3.49INDD395 pKa = 3.32VEE397 pKa = 5.22IITDD401 pKa = 3.37YY402 pKa = 11.59AGIVNSEE409 pKa = 3.3NDD411 pKa = 3.5YY412 pKa = 11.57NVDD415 pKa = 3.34FTTAVRR421 pKa = 11.84VNVQNLLSEE430 pKa = 4.39NLPVSTSMLFSSLDD444 pKa = 3.32YY445 pKa = 10.88AKK447 pKa = 10.67NYY449 pKa = 10.01GFIGFSPLTGNEE461 pKa = 3.57IDD463 pKa = 4.34CYY465 pKa = 11.34GDD467 pKa = 3.13ILFDD471 pKa = 3.61IFAGDD476 pKa = 3.85KK477 pKa = 11.29YY478 pKa = 11.08NICSDD483 pKa = 3.91NEE485 pKa = 4.04ISSSNAIDD493 pKa = 4.02DD494 pKa = 4.24NEE496 pKa = 4.13FFSTYY501 pKa = 8.04TKK503 pKa = 11.03NVGTTTLTSPCRR515 pKa = 11.84VKK517 pKa = 10.98SDD519 pKa = 3.15IVLEE523 pKa = 4.04DD524 pKa = 4.88DD525 pKa = 4.88IYY527 pKa = 10.99IQEE530 pKa = 4.14SDD532 pKa = 3.74NVSVSCLYY540 pKa = 11.18AMPLQEE546 pKa = 5.9IIVTDD551 pKa = 4.04LEE553 pKa = 4.13EE554 pKa = 4.18AAGVLIDD561 pKa = 5.45DD562 pKa = 4.45KK563 pKa = 11.29CSEE566 pKa = 4.45NGIDD570 pKa = 3.87LNNVAGAIVILNDD583 pKa = 3.2EE584 pKa = 4.31LACRR588 pKa = 11.84SVYY591 pKa = 10.89LEE593 pKa = 4.92DD594 pKa = 3.72YY595 pKa = 10.71NVIITASFTPTMKK608 pKa = 10.76FEE610 pKa = 4.59DD611 pKa = 3.96RR612 pKa = 11.84TWNVDD617 pKa = 3.19TQSYY621 pKa = 10.09DD622 pKa = 2.77GVMYY626 pKa = 10.53DD627 pKa = 3.92SVVHH631 pKa = 6.89KK632 pKa = 10.23ITDD635 pKa = 2.99GWMACPPSAYY645 pKa = 10.33DD646 pKa = 4.93KK647 pKa = 8.26TTTDD651 pKa = 3.98CNIDD655 pKa = 3.36NTKK658 pKa = 10.77LYY660 pKa = 9.11TVKK663 pKa = 10.61DD664 pKa = 3.16LHH666 pKa = 6.65FVQNYY671 pKa = 9.27PDD673 pKa = 3.61IDD675 pKa = 3.91YY676 pKa = 11.07EE677 pKa = 4.21PEE679 pKa = 3.81NEE681 pKa = 4.26VNVDD685 pKa = 3.61TEE687 pKa = 4.8DD688 pKa = 3.75VDD690 pKa = 3.98VSNPIADD697 pKa = 3.88DD698 pKa = 3.82VEE700 pKa = 5.29AINPIAEE707 pKa = 4.22DD708 pKa = 3.52VLRR711 pKa = 11.84QNDD714 pKa = 3.46NVTNNVQPALEE725 pKa = 4.23NEE727 pKa = 4.6LVNTATQMEE736 pKa = 4.66VDD738 pKa = 3.87QKK740 pKa = 11.24NPSINVQDD748 pKa = 3.92IDD750 pKa = 4.18SSSTLSDD757 pKa = 3.06EE758 pKa = 4.54SVKK761 pKa = 10.78NDD763 pKa = 3.41YY764 pKa = 9.11STLDD768 pKa = 3.28TNRR771 pKa = 11.84ILDD774 pKa = 4.06YY775 pKa = 10.77INRR778 pKa = 11.84HH779 pKa = 5.78HH780 pKa = 6.86EE781 pKa = 4.28SEE783 pKa = 4.36TSEE786 pKa = 4.28EE787 pKa = 4.46EE788 pKa = 4.14YY789 pKa = 10.48MDD791 pKa = 5.57DD792 pKa = 4.73GDD794 pKa = 4.04ILASLFEE801 pKa = 4.25
Molecular weight: 90.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q0ZNZ6|Q0ZNZ6_9CBAC Uncharacterized protein OS=Neodiprion abietis NPV OX=204507 PE=4 SV=1
MM1 pKa = 7.53LSNRR5 pKa = 11.84ITAEE9 pKa = 3.78TIHH12 pKa = 7.34RR13 pKa = 11.84YY14 pKa = 9.26LCYY17 pKa = 10.45SNNYY21 pKa = 8.69RR22 pKa = 11.84LIVSAVKK29 pKa = 10.12RR30 pKa = 11.84SSEE33 pKa = 3.75ISVSSVTRR41 pKa = 11.84IIYY44 pKa = 8.82AQKK47 pKa = 10.9KK48 pKa = 9.37LVLFALL54 pKa = 4.49
MM1 pKa = 7.53LSNRR5 pKa = 11.84ITAEE9 pKa = 3.78TIHH12 pKa = 7.34RR13 pKa = 11.84YY14 pKa = 9.26LCYY17 pKa = 10.45SNNYY21 pKa = 8.69RR22 pKa = 11.84LIVSAVKK29 pKa = 10.12RR30 pKa = 11.84SSEE33 pKa = 3.75ISVSSVTRR41 pKa = 11.84IIYY44 pKa = 8.82AQKK47 pKa = 10.9KK48 pKa = 9.37LVLFALL54 pKa = 4.49
Molecular weight: 6.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
24792 |
51 |
1136 |
266.6 |
30.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.574 ± 0.194 | 2.686 ± 0.171 |
6.599 ± 0.212 | 5.203 ± 0.226 |
4.852 ± 0.177 | 2.836 ± 0.173 |
2.339 ± 0.126 | 8.704 ± 0.184 |
7.176 ± 0.301 | 9.076 ± 0.208 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.424 ± 0.1 | 8.132 ± 0.208 |
2.775 ± 0.158 | 3.562 ± 0.141 |
4.013 ± 0.194 | 6.877 ± 0.23 |
6.954 ± 0.247 | 6.381 ± 0.203 |
0.811 ± 0.075 | 5.026 ± 0.162 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
