
Lake Sarah-associated circular molecule 2
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.24
Get precalculated fractions of proteins
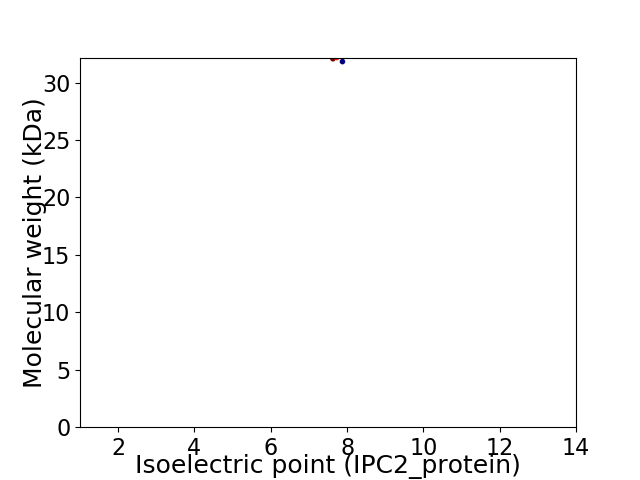
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126G9F6|A0A126G9F6_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular molecule 2 OX=1685727 PE=3 SV=1
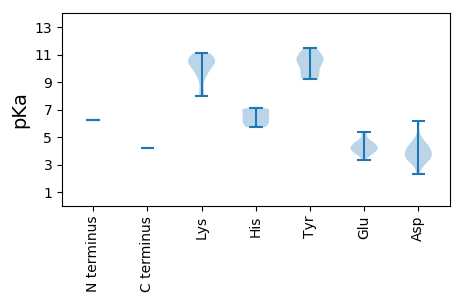
MM1 pKa = 6.27STRR4 pKa = 11.84RR5 pKa = 11.84IVFTKK10 pKa = 10.85NNWTEE15 pKa = 3.35EE16 pKa = 4.2DD17 pKa = 3.54YY18 pKa = 10.92RR19 pKa = 11.84ALMEE23 pKa = 4.7EE24 pKa = 4.39PLFSYY29 pKa = 10.11LIIGKK34 pKa = 9.6EE35 pKa = 3.76IGEE38 pKa = 4.53KK39 pKa = 9.31GTPHH43 pKa = 5.85LQGYY47 pKa = 10.53AEE49 pKa = 4.08FTKK52 pKa = 10.26KK53 pKa = 10.53AKK55 pKa = 10.55YY56 pKa = 9.33GALAKK61 pKa = 9.83KK62 pKa = 10.36YY63 pKa = 10.45KK64 pKa = 8.33MHH66 pKa = 6.98CEE68 pKa = 4.11VSRR71 pKa = 11.84GTQDD75 pKa = 3.26EE76 pKa = 4.52AIKK79 pKa = 11.06YY80 pKa = 9.22CMKK83 pKa = 10.91DD84 pKa = 2.34GDD86 pKa = 4.12YY87 pKa = 11.42AEE89 pKa = 5.36KK90 pKa = 9.67GTKK93 pKa = 9.99KK94 pKa = 10.56LDD96 pKa = 3.52GASAEE101 pKa = 4.23KK102 pKa = 10.65NRR104 pKa = 11.84WEE106 pKa = 4.05QARR109 pKa = 11.84VSAKK113 pKa = 10.02EE114 pKa = 3.74GRR116 pKa = 11.84LDD118 pKa = 4.74DD119 pKa = 4.18VPADD123 pKa = 3.34IYY125 pKa = 10.5IRR127 pKa = 11.84CYY129 pKa = 9.69RR130 pKa = 11.84TLKK133 pKa = 10.65EE134 pKa = 3.75IAKK137 pKa = 10.64DD138 pKa = 3.61NMPKK142 pKa = 10.24PEE144 pKa = 4.58SLTEE148 pKa = 4.49LKK150 pKa = 10.76NLWIYY155 pKa = 10.85GPAGSGKK162 pKa = 8.01TRR164 pKa = 11.84LADD167 pKa = 4.44AIVPISYY174 pKa = 10.61SKK176 pKa = 11.09NCNKK180 pKa = 9.25WWDD183 pKa = 4.07GYY185 pKa = 9.2QNEE188 pKa = 4.76PGVIINDD195 pKa = 3.35VGKK198 pKa = 8.2EE199 pKa = 3.83HH200 pKa = 6.96SVLGHH205 pKa = 6.24HH206 pKa = 6.58FKK208 pKa = 10.96LWGEE212 pKa = 3.96HH213 pKa = 5.72RR214 pKa = 11.84PFIAEE219 pKa = 4.08TKK221 pKa = 10.27GGAIHH226 pKa = 7.08IRR228 pKa = 11.84PQRR231 pKa = 11.84VIITSQYY238 pKa = 10.87SLTQIWEE245 pKa = 4.2DD246 pKa = 3.73EE247 pKa = 4.24EE248 pKa = 4.42TRR250 pKa = 11.84DD251 pKa = 4.2ALARR255 pKa = 11.84RR256 pKa = 11.84YY257 pKa = 9.61KK258 pKa = 10.44VLHH261 pKa = 6.17LLGNLDD267 pKa = 4.57LEE269 pKa = 5.2DD270 pKa = 3.87IDD272 pKa = 6.16RR273 pKa = 11.84ISKK276 pKa = 10.54DD277 pKa = 3.43YY278 pKa = 11.45AA279 pKa = 4.18
MM1 pKa = 6.27STRR4 pKa = 11.84RR5 pKa = 11.84IVFTKK10 pKa = 10.85NNWTEE15 pKa = 3.35EE16 pKa = 4.2DD17 pKa = 3.54YY18 pKa = 10.92RR19 pKa = 11.84ALMEE23 pKa = 4.7EE24 pKa = 4.39PLFSYY29 pKa = 10.11LIIGKK34 pKa = 9.6EE35 pKa = 3.76IGEE38 pKa = 4.53KK39 pKa = 9.31GTPHH43 pKa = 5.85LQGYY47 pKa = 10.53AEE49 pKa = 4.08FTKK52 pKa = 10.26KK53 pKa = 10.53AKK55 pKa = 10.55YY56 pKa = 9.33GALAKK61 pKa = 9.83KK62 pKa = 10.36YY63 pKa = 10.45KK64 pKa = 8.33MHH66 pKa = 6.98CEE68 pKa = 4.11VSRR71 pKa = 11.84GTQDD75 pKa = 3.26EE76 pKa = 4.52AIKK79 pKa = 11.06YY80 pKa = 9.22CMKK83 pKa = 10.91DD84 pKa = 2.34GDD86 pKa = 4.12YY87 pKa = 11.42AEE89 pKa = 5.36KK90 pKa = 9.67GTKK93 pKa = 9.99KK94 pKa = 10.56LDD96 pKa = 3.52GASAEE101 pKa = 4.23KK102 pKa = 10.65NRR104 pKa = 11.84WEE106 pKa = 4.05QARR109 pKa = 11.84VSAKK113 pKa = 10.02EE114 pKa = 3.74GRR116 pKa = 11.84LDD118 pKa = 4.74DD119 pKa = 4.18VPADD123 pKa = 3.34IYY125 pKa = 10.5IRR127 pKa = 11.84CYY129 pKa = 9.69RR130 pKa = 11.84TLKK133 pKa = 10.65EE134 pKa = 3.75IAKK137 pKa = 10.64DD138 pKa = 3.61NMPKK142 pKa = 10.24PEE144 pKa = 4.58SLTEE148 pKa = 4.49LKK150 pKa = 10.76NLWIYY155 pKa = 10.85GPAGSGKK162 pKa = 8.01TRR164 pKa = 11.84LADD167 pKa = 4.44AIVPISYY174 pKa = 10.61SKK176 pKa = 11.09NCNKK180 pKa = 9.25WWDD183 pKa = 4.07GYY185 pKa = 9.2QNEE188 pKa = 4.76PGVIINDD195 pKa = 3.35VGKK198 pKa = 8.2EE199 pKa = 3.83HH200 pKa = 6.96SVLGHH205 pKa = 6.24HH206 pKa = 6.58FKK208 pKa = 10.96LWGEE212 pKa = 3.96HH213 pKa = 5.72RR214 pKa = 11.84PFIAEE219 pKa = 4.08TKK221 pKa = 10.27GGAIHH226 pKa = 7.08IRR228 pKa = 11.84PQRR231 pKa = 11.84VIITSQYY238 pKa = 10.87SLTQIWEE245 pKa = 4.2DD246 pKa = 3.73EE247 pKa = 4.24EE248 pKa = 4.42TRR250 pKa = 11.84DD251 pKa = 4.2ALARR255 pKa = 11.84RR256 pKa = 11.84YY257 pKa = 9.61KK258 pKa = 10.44VLHH261 pKa = 6.17LLGNLDD267 pKa = 4.57LEE269 pKa = 5.2DD270 pKa = 3.87IDD272 pKa = 6.16RR273 pKa = 11.84ISKK276 pKa = 10.54DD277 pKa = 3.43YY278 pKa = 11.45AA279 pKa = 4.18
Molecular weight: 32.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126G9F6|A0A126G9F6_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular molecule 2 OX=1685727 PE=3 SV=1
MM1 pKa = 6.27STRR4 pKa = 11.84RR5 pKa = 11.84IVFTKK10 pKa = 10.85NNWTEE15 pKa = 3.35EE16 pKa = 4.2DD17 pKa = 3.54YY18 pKa = 10.92RR19 pKa = 11.84ALMEE23 pKa = 4.7EE24 pKa = 4.39PLFSYY29 pKa = 10.11LIIGKK34 pKa = 9.6EE35 pKa = 3.76IGEE38 pKa = 4.53KK39 pKa = 9.31GTPHH43 pKa = 5.85LQGYY47 pKa = 10.53AEE49 pKa = 4.08FTKK52 pKa = 10.26KK53 pKa = 10.53AKK55 pKa = 10.55YY56 pKa = 9.33GALAKK61 pKa = 9.83KK62 pKa = 10.36YY63 pKa = 10.45KK64 pKa = 8.33MHH66 pKa = 6.98CEE68 pKa = 4.11VSRR71 pKa = 11.84GTQDD75 pKa = 3.26EE76 pKa = 4.52AIKK79 pKa = 11.06YY80 pKa = 9.22CMKK83 pKa = 10.91DD84 pKa = 2.34GDD86 pKa = 4.12YY87 pKa = 11.42AEE89 pKa = 5.36KK90 pKa = 9.67GTKK93 pKa = 9.99KK94 pKa = 10.56LDD96 pKa = 3.52GASAEE101 pKa = 4.23KK102 pKa = 10.65NRR104 pKa = 11.84WEE106 pKa = 4.05QARR109 pKa = 11.84VSAKK113 pKa = 10.02EE114 pKa = 3.74GRR116 pKa = 11.84LDD118 pKa = 4.74DD119 pKa = 4.18VPADD123 pKa = 3.34IYY125 pKa = 10.5IRR127 pKa = 11.84CYY129 pKa = 9.69RR130 pKa = 11.84TLKK133 pKa = 10.65EE134 pKa = 3.75IAKK137 pKa = 10.64DD138 pKa = 3.61NMPKK142 pKa = 10.24PEE144 pKa = 4.58SLTEE148 pKa = 4.49LKK150 pKa = 10.76NLWIYY155 pKa = 10.85GPAGSGKK162 pKa = 8.01TRR164 pKa = 11.84LADD167 pKa = 4.44AIVPISYY174 pKa = 10.61SKK176 pKa = 11.09NCNKK180 pKa = 9.25WWDD183 pKa = 4.07GYY185 pKa = 9.2QNEE188 pKa = 4.76PGVIINDD195 pKa = 3.35VGKK198 pKa = 8.2EE199 pKa = 3.83HH200 pKa = 6.96SVLGHH205 pKa = 6.24HH206 pKa = 6.58FKK208 pKa = 10.96LWGEE212 pKa = 3.96HH213 pKa = 5.72RR214 pKa = 11.84PFIAEE219 pKa = 4.08TKK221 pKa = 10.27GGAIHH226 pKa = 7.08IRR228 pKa = 11.84PQRR231 pKa = 11.84VIITSQYY238 pKa = 10.87SLTQIWEE245 pKa = 4.2DD246 pKa = 3.73EE247 pKa = 4.24EE248 pKa = 4.42TRR250 pKa = 11.84DD251 pKa = 4.2ALARR255 pKa = 11.84RR256 pKa = 11.84YY257 pKa = 9.61KK258 pKa = 10.44VLHH261 pKa = 6.17LLGNLDD267 pKa = 4.57LEE269 pKa = 5.2DD270 pKa = 3.87IDD272 pKa = 6.16RR273 pKa = 11.84ISKK276 pKa = 10.54DD277 pKa = 3.43YY278 pKa = 11.45AA279 pKa = 4.18
MM1 pKa = 6.27STRR4 pKa = 11.84RR5 pKa = 11.84IVFTKK10 pKa = 10.85NNWTEE15 pKa = 3.35EE16 pKa = 4.2DD17 pKa = 3.54YY18 pKa = 10.92RR19 pKa = 11.84ALMEE23 pKa = 4.7EE24 pKa = 4.39PLFSYY29 pKa = 10.11LIIGKK34 pKa = 9.6EE35 pKa = 3.76IGEE38 pKa = 4.53KK39 pKa = 9.31GTPHH43 pKa = 5.85LQGYY47 pKa = 10.53AEE49 pKa = 4.08FTKK52 pKa = 10.26KK53 pKa = 10.53AKK55 pKa = 10.55YY56 pKa = 9.33GALAKK61 pKa = 9.83KK62 pKa = 10.36YY63 pKa = 10.45KK64 pKa = 8.33MHH66 pKa = 6.98CEE68 pKa = 4.11VSRR71 pKa = 11.84GTQDD75 pKa = 3.26EE76 pKa = 4.52AIKK79 pKa = 11.06YY80 pKa = 9.22CMKK83 pKa = 10.91DD84 pKa = 2.34GDD86 pKa = 4.12YY87 pKa = 11.42AEE89 pKa = 5.36KK90 pKa = 9.67GTKK93 pKa = 9.99KK94 pKa = 10.56LDD96 pKa = 3.52GASAEE101 pKa = 4.23KK102 pKa = 10.65NRR104 pKa = 11.84WEE106 pKa = 4.05QARR109 pKa = 11.84VSAKK113 pKa = 10.02EE114 pKa = 3.74GRR116 pKa = 11.84LDD118 pKa = 4.74DD119 pKa = 4.18VPADD123 pKa = 3.34IYY125 pKa = 10.5IRR127 pKa = 11.84CYY129 pKa = 9.69RR130 pKa = 11.84TLKK133 pKa = 10.65EE134 pKa = 3.75IAKK137 pKa = 10.64DD138 pKa = 3.61NMPKK142 pKa = 10.24PEE144 pKa = 4.58SLTEE148 pKa = 4.49LKK150 pKa = 10.76NLWIYY155 pKa = 10.85GPAGSGKK162 pKa = 8.01TRR164 pKa = 11.84LADD167 pKa = 4.44AIVPISYY174 pKa = 10.61SKK176 pKa = 11.09NCNKK180 pKa = 9.25WWDD183 pKa = 4.07GYY185 pKa = 9.2QNEE188 pKa = 4.76PGVIINDD195 pKa = 3.35VGKK198 pKa = 8.2EE199 pKa = 3.83HH200 pKa = 6.96SVLGHH205 pKa = 6.24HH206 pKa = 6.58FKK208 pKa = 10.96LWGEE212 pKa = 3.96HH213 pKa = 5.72RR214 pKa = 11.84PFIAEE219 pKa = 4.08TKK221 pKa = 10.27GGAIHH226 pKa = 7.08IRR228 pKa = 11.84PQRR231 pKa = 11.84VIITSQYY238 pKa = 10.87SLTQIWEE245 pKa = 4.2DD246 pKa = 3.73EE247 pKa = 4.24EE248 pKa = 4.42TRR250 pKa = 11.84DD251 pKa = 4.2ALARR255 pKa = 11.84RR256 pKa = 11.84YY257 pKa = 9.61KK258 pKa = 10.44VLHH261 pKa = 6.17LLGNLDD267 pKa = 4.57LEE269 pKa = 5.2DD270 pKa = 3.87IDD272 pKa = 6.16RR273 pKa = 11.84ISKK276 pKa = 10.54DD277 pKa = 3.43YY278 pKa = 11.45AA279 pKa = 4.18
Molecular weight: 32.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
279 |
279 |
279 |
279.0 |
32.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.527 ± 0.0 | 1.434 ± 0.0 |
6.452 ± 0.0 | 8.602 ± 0.0 |
1.792 ± 0.0 | 7.527 ± 0.0 |
2.867 ± 0.0 | 7.527 ± 0.0 |
10.036 ± 0.0 | 7.527 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.792 ± 0.0 | 3.584 ± 0.0 |
3.584 ± 0.0 | 2.509 ± 0.0 |
6.093 ± 0.0 | 4.659 ± 0.0 |
5.018 ± 0.0 | 3.584 ± 0.0 |
2.509 ± 0.0 | 5.376 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
