
Diplodia corticola
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetes incertae sedis; Botryosphaeriales; Botryosphaeriaceae; Diplodia
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
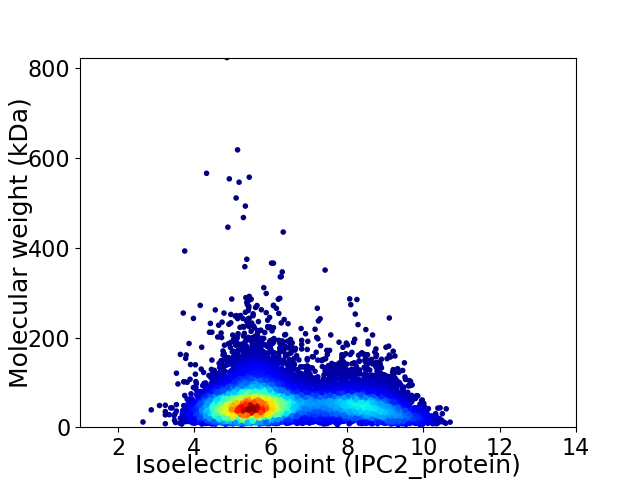
Virtual 2D-PAGE plot for 10836 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1J9RLD5|A0A1J9RLD5_9PEZI Ste ste20 ysk protein kinase OS=Diplodia corticola OX=236234 GN=BKCO1_3100084 PE=4 SV=1
MM1 pKa = 7.4SPHH4 pKa = 6.95LSFLSATLAVFPFWVSPVSTIALSSLKK31 pKa = 10.77NEE33 pKa = 4.18STPAAAHH40 pKa = 6.57LDD42 pKa = 3.35IATSSGTTVSSTPPAQLAASITPTPANSTTQHH74 pKa = 6.68DD75 pKa = 4.27ASDD78 pKa = 3.57AAALSRR84 pKa = 11.84NRR86 pKa = 11.84ACEE89 pKa = 3.9SLIQSWNSASASFAASILAGSPDD112 pKa = 3.49AAASALATIPHH123 pKa = 6.01SLITTTYY130 pKa = 8.28PGGYY134 pKa = 8.45TISTSYY140 pKa = 11.35NPLLLPHH147 pKa = 6.25FTSTPSAPSTPSPNATISPQPSASVSTPCADD178 pKa = 5.0RR179 pKa = 11.84IPRR182 pKa = 11.84APGAAYY188 pKa = 10.69SLLTVATASPTTLVSTRR205 pKa = 11.84IAYY208 pKa = 7.69EE209 pKa = 3.84ALVPAPACAYY219 pKa = 10.02DD220 pKa = 3.67RR221 pKa = 11.84RR222 pKa = 11.84GCALEE227 pKa = 4.51LAEE230 pKa = 5.25KK231 pKa = 10.41PKK233 pKa = 9.95TLPLCAQGGNVAAGFVNDD251 pKa = 4.5EE252 pKa = 4.35KK253 pKa = 11.22CVLPSGVVGCRR264 pKa = 11.84VAAGGVGVYY273 pKa = 7.83YY274 pKa = 10.12WPRR277 pKa = 11.84EE278 pKa = 4.04DD279 pKa = 4.34GGEE282 pKa = 4.24GACAGGGEE290 pKa = 4.55GEE292 pKa = 4.57GEE294 pKa = 4.41GEE296 pKa = 4.29GKK298 pKa = 10.62VGVSAAPTTTKK309 pKa = 7.45TTKK312 pKa = 9.77TVVVGTNTFTSPTVYY327 pKa = 10.66VSFSEE332 pKa = 3.98IDD334 pKa = 3.19AAPYY338 pKa = 9.83CLPTYY343 pKa = 10.25RR344 pKa = 11.84NVVVGLPPEE353 pKa = 4.38SVSWVSMSRR362 pKa = 11.84GWPITKK368 pKa = 10.08SLNWNDD374 pKa = 3.35LNYY377 pKa = 9.68PVPSTAYY384 pKa = 9.92HH385 pKa = 6.24GMEE388 pKa = 3.88ACQVNTAALRR398 pKa = 11.84PDD400 pKa = 3.74GCATIYY406 pKa = 11.13DD407 pKa = 4.38DD408 pKa = 6.35DD409 pKa = 4.13YY410 pKa = 11.63HH411 pKa = 7.23PYY413 pKa = 10.37LALPTDD419 pKa = 3.65PALYY423 pKa = 9.69QQALGGSDD431 pKa = 3.18GGGGFANCTALTWQKK446 pKa = 11.02YY447 pKa = 6.55VTDD450 pKa = 4.31PPIPLTTAGALIPQSSEE467 pKa = 3.63VPAATTTTTTTTGADD482 pKa = 3.74PEE484 pKa = 4.72STTAAAPVTEE494 pKa = 4.77TNAPTAPATATATTGGGGGNNAQATGSSSAGSSDD528 pKa = 3.54PADD531 pKa = 4.0PSATSAVVDD540 pKa = 3.73TNPWDD545 pKa = 3.72NLGSMISEE553 pKa = 4.36AWSTSFADD561 pKa = 4.13PAAEE565 pKa = 4.07TATTTTGTAAGQGGGGEE582 pKa = 4.36ASDD585 pKa = 3.8GLPVVEE591 pKa = 5.22TSAPASSAPEE601 pKa = 4.25PPSSAAVVATIGTAVASPAAPASAADD627 pKa = 3.87SSDD630 pKa = 3.24PTATATAGNNNDD642 pKa = 4.17NNDD645 pKa = 4.74DD646 pKa = 4.0DD647 pKa = 6.1DD648 pKa = 4.83EE649 pKa = 5.14NNNSNSNNSDD659 pKa = 3.39NNDD662 pKa = 3.33NGSPGGSQTLPAGHH676 pKa = 6.18TTATAPLYY684 pKa = 10.2TPTASSSPAPASPAPPIAITLSDD707 pKa = 3.98DD708 pKa = 3.62SGSGSGSGSGSHH720 pKa = 7.07VIAADD725 pKa = 3.72PSDD728 pKa = 3.63PTAGVIVVGGGAGGDD743 pKa = 3.73AQEE746 pKa = 4.91DD747 pKa = 4.16ASTQTIAAGATTTIDD762 pKa = 3.2GTQVVVSSVPAAEE775 pKa = 4.67GDD777 pKa = 3.74SDD779 pKa = 5.85GDD781 pKa = 5.23DD782 pKa = 4.96SDD784 pKa = 6.6DD785 pKa = 6.38DD786 pKa = 6.47DD787 pKa = 7.7DD788 pKa = 7.64DD789 pKa = 7.65DD790 pKa = 7.68DD791 pKa = 7.68DD792 pKa = 7.63DD793 pKa = 7.58DD794 pKa = 7.46DD795 pKa = 7.79DD796 pKa = 5.46DD797 pKa = 4.57AQPPAVVVVVVGGSRR812 pKa = 11.84TYY814 pKa = 9.91TLPGGATTGGSGSGSGSGGGIVGATTVPLSSLAAGTAGGAGRR856 pKa = 11.84TGAVVTFTAGGAGGGGEE873 pKa = 4.27KK874 pKa = 9.69TLSAAQLAPGTVVVGGKK891 pKa = 7.04TLSAGGPAATVTVSGTGSGLGSGLSGDD918 pKa = 3.97GEE920 pKa = 4.85VVVVASVAPGGGLVVGSAGGGGGGSSSTSRR950 pKa = 11.84RR951 pKa = 11.84LSSAAATASRR961 pKa = 11.84TMSADD966 pKa = 2.87NTATRR971 pKa = 11.84NSALPTTTATATTPDD986 pKa = 5.02DD987 pKa = 5.03DD988 pKa = 7.05DD989 pKa = 6.33SDD991 pKa = 5.38NDD993 pKa = 5.03DD994 pKa = 5.75DD995 pKa = 6.99DD996 pKa = 6.86DD997 pKa = 6.02DD998 pKa = 4.6SQSTDD1003 pKa = 2.91AAAAAATATTAGAASGGSVPPLLAMLCSFLLTCSIPSVFSIFAA1046 pKa = 4.02
MM1 pKa = 7.4SPHH4 pKa = 6.95LSFLSATLAVFPFWVSPVSTIALSSLKK31 pKa = 10.77NEE33 pKa = 4.18STPAAAHH40 pKa = 6.57LDD42 pKa = 3.35IATSSGTTVSSTPPAQLAASITPTPANSTTQHH74 pKa = 6.68DD75 pKa = 4.27ASDD78 pKa = 3.57AAALSRR84 pKa = 11.84NRR86 pKa = 11.84ACEE89 pKa = 3.9SLIQSWNSASASFAASILAGSPDD112 pKa = 3.49AAASALATIPHH123 pKa = 6.01SLITTTYY130 pKa = 8.28PGGYY134 pKa = 8.45TISTSYY140 pKa = 11.35NPLLLPHH147 pKa = 6.25FTSTPSAPSTPSPNATISPQPSASVSTPCADD178 pKa = 5.0RR179 pKa = 11.84IPRR182 pKa = 11.84APGAAYY188 pKa = 10.69SLLTVATASPTTLVSTRR205 pKa = 11.84IAYY208 pKa = 7.69EE209 pKa = 3.84ALVPAPACAYY219 pKa = 10.02DD220 pKa = 3.67RR221 pKa = 11.84RR222 pKa = 11.84GCALEE227 pKa = 4.51LAEE230 pKa = 5.25KK231 pKa = 10.41PKK233 pKa = 9.95TLPLCAQGGNVAAGFVNDD251 pKa = 4.5EE252 pKa = 4.35KK253 pKa = 11.22CVLPSGVVGCRR264 pKa = 11.84VAAGGVGVYY273 pKa = 7.83YY274 pKa = 10.12WPRR277 pKa = 11.84EE278 pKa = 4.04DD279 pKa = 4.34GGEE282 pKa = 4.24GACAGGGEE290 pKa = 4.55GEE292 pKa = 4.57GEE294 pKa = 4.41GEE296 pKa = 4.29GKK298 pKa = 10.62VGVSAAPTTTKK309 pKa = 7.45TTKK312 pKa = 9.77TVVVGTNTFTSPTVYY327 pKa = 10.66VSFSEE332 pKa = 3.98IDD334 pKa = 3.19AAPYY338 pKa = 9.83CLPTYY343 pKa = 10.25RR344 pKa = 11.84NVVVGLPPEE353 pKa = 4.38SVSWVSMSRR362 pKa = 11.84GWPITKK368 pKa = 10.08SLNWNDD374 pKa = 3.35LNYY377 pKa = 9.68PVPSTAYY384 pKa = 9.92HH385 pKa = 6.24GMEE388 pKa = 3.88ACQVNTAALRR398 pKa = 11.84PDD400 pKa = 3.74GCATIYY406 pKa = 11.13DD407 pKa = 4.38DD408 pKa = 6.35DD409 pKa = 4.13YY410 pKa = 11.63HH411 pKa = 7.23PYY413 pKa = 10.37LALPTDD419 pKa = 3.65PALYY423 pKa = 9.69QQALGGSDD431 pKa = 3.18GGGGFANCTALTWQKK446 pKa = 11.02YY447 pKa = 6.55VTDD450 pKa = 4.31PPIPLTTAGALIPQSSEE467 pKa = 3.63VPAATTTTTTTTGADD482 pKa = 3.74PEE484 pKa = 4.72STTAAAPVTEE494 pKa = 4.77TNAPTAPATATATTGGGGGNNAQATGSSSAGSSDD528 pKa = 3.54PADD531 pKa = 4.0PSATSAVVDD540 pKa = 3.73TNPWDD545 pKa = 3.72NLGSMISEE553 pKa = 4.36AWSTSFADD561 pKa = 4.13PAAEE565 pKa = 4.07TATTTTGTAAGQGGGGEE582 pKa = 4.36ASDD585 pKa = 3.8GLPVVEE591 pKa = 5.22TSAPASSAPEE601 pKa = 4.25PPSSAAVVATIGTAVASPAAPASAADD627 pKa = 3.87SSDD630 pKa = 3.24PTATATAGNNNDD642 pKa = 4.17NNDD645 pKa = 4.74DD646 pKa = 4.0DD647 pKa = 6.1DD648 pKa = 4.83EE649 pKa = 5.14NNNSNSNNSDD659 pKa = 3.39NNDD662 pKa = 3.33NGSPGGSQTLPAGHH676 pKa = 6.18TTATAPLYY684 pKa = 10.2TPTASSSPAPASPAPPIAITLSDD707 pKa = 3.98DD708 pKa = 3.62SGSGSGSGSGSHH720 pKa = 7.07VIAADD725 pKa = 3.72PSDD728 pKa = 3.63PTAGVIVVGGGAGGDD743 pKa = 3.73AQEE746 pKa = 4.91DD747 pKa = 4.16ASTQTIAAGATTTIDD762 pKa = 3.2GTQVVVSSVPAAEE775 pKa = 4.67GDD777 pKa = 3.74SDD779 pKa = 5.85GDD781 pKa = 5.23DD782 pKa = 4.96SDD784 pKa = 6.6DD785 pKa = 6.38DD786 pKa = 6.47DD787 pKa = 7.7DD788 pKa = 7.64DD789 pKa = 7.65DD790 pKa = 7.68DD791 pKa = 7.68DD792 pKa = 7.63DD793 pKa = 7.58DD794 pKa = 7.46DD795 pKa = 7.79DD796 pKa = 5.46DD797 pKa = 4.57AQPPAVVVVVVGGSRR812 pKa = 11.84TYY814 pKa = 9.91TLPGGATTGGSGSGSGSGGGIVGATTVPLSSLAAGTAGGAGRR856 pKa = 11.84TGAVVTFTAGGAGGGGEE873 pKa = 4.27KK874 pKa = 9.69TLSAAQLAPGTVVVGGKK891 pKa = 7.04TLSAGGPAATVTVSGTGSGLGSGLSGDD918 pKa = 3.97GEE920 pKa = 4.85VVVVASVAPGGGLVVGSAGGGGGGSSSTSRR950 pKa = 11.84RR951 pKa = 11.84LSSAAATASRR961 pKa = 11.84TMSADD966 pKa = 2.87NTATRR971 pKa = 11.84NSALPTTTATATTPDD986 pKa = 5.02DD987 pKa = 5.03DD988 pKa = 7.05DD989 pKa = 6.33SDD991 pKa = 5.38NDD993 pKa = 5.03DD994 pKa = 5.75DD995 pKa = 6.99DD996 pKa = 6.86DD997 pKa = 6.02DD998 pKa = 4.6SQSTDD1003 pKa = 2.91AAAAAATATTAGAASGGSVPPLLAMLCSFLLTCSIPSVFSIFAA1046 pKa = 4.02
Molecular weight: 101.35 kDa
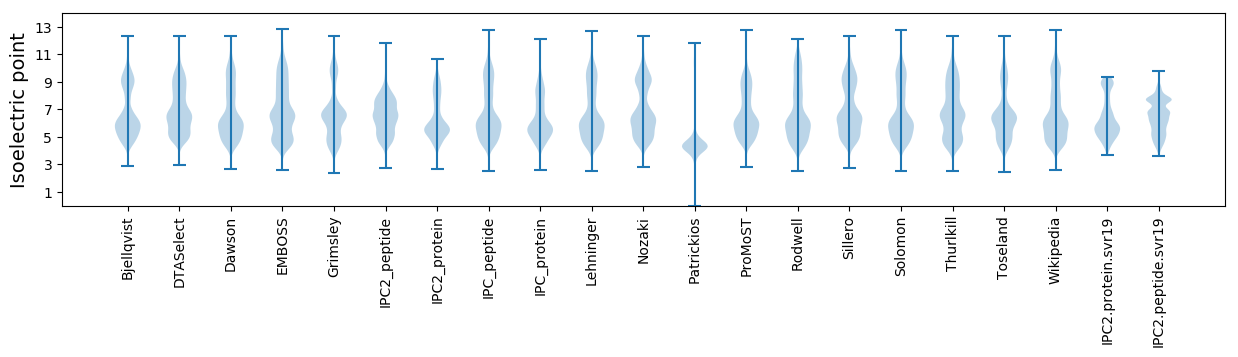
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1J9R6V3|A0A1J9R6V3_9PEZI Branchpoint-bridging protein OS=Diplodia corticola OX=236234 GN=BKCO1_12000130 PE=3 SV=1
MM1 pKa = 7.34ATKK4 pKa = 10.4QSTKK8 pKa = 9.15YY9 pKa = 10.05EE10 pKa = 4.05RR11 pKa = 11.84LEE13 pKa = 4.08GLSQRR18 pKa = 11.84RR19 pKa = 11.84GVDD22 pKa = 3.3TQHH25 pKa = 6.39AQQAEE30 pKa = 3.94RR31 pKa = 11.84DD32 pKa = 3.68RR33 pKa = 11.84RR34 pKa = 11.84LRR36 pKa = 11.84WAQEE40 pKa = 3.87MIMQNPALCAEE51 pKa = 5.15VYY53 pKa = 10.47CAMSKK58 pKa = 10.28RR59 pKa = 11.84PAMPPRR65 pKa = 11.84RR66 pKa = 11.84EE67 pKa = 4.01SSLSSSSSSSRR78 pKa = 11.84RR79 pKa = 11.84SPSTAAGSMPSPGISSNSSINSGSPTT105 pKa = 3.32
MM1 pKa = 7.34ATKK4 pKa = 10.4QSTKK8 pKa = 9.15YY9 pKa = 10.05EE10 pKa = 4.05RR11 pKa = 11.84LEE13 pKa = 4.08GLSQRR18 pKa = 11.84RR19 pKa = 11.84GVDD22 pKa = 3.3TQHH25 pKa = 6.39AQQAEE30 pKa = 3.94RR31 pKa = 11.84DD32 pKa = 3.68RR33 pKa = 11.84RR34 pKa = 11.84LRR36 pKa = 11.84WAQEE40 pKa = 3.87MIMQNPALCAEE51 pKa = 5.15VYY53 pKa = 10.47CAMSKK58 pKa = 10.28RR59 pKa = 11.84PAMPPRR65 pKa = 11.84RR66 pKa = 11.84EE67 pKa = 4.01SSLSSSSSSSRR78 pKa = 11.84RR79 pKa = 11.84SPSTAAGSMPSPGISSNSSINSGSPTT105 pKa = 3.32
Molecular weight: 11.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5743668 |
55 |
7609 |
530.1 |
58.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.894 ± 0.026 | 1.162 ± 0.009 |
5.859 ± 0.02 | 6.145 ± 0.024 |
3.52 ± 0.015 | 7.395 ± 0.028 |
2.432 ± 0.011 | 4.136 ± 0.016 |
4.472 ± 0.018 | 8.523 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.067 ± 0.008 | 3.39 ± 0.013 |
6.43 ± 0.033 | 3.901 ± 0.019 |
6.345 ± 0.023 | 8.097 ± 0.03 |
5.946 ± 0.024 | 6.227 ± 0.018 |
1.464 ± 0.009 | 2.592 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
