
Treponema pallidum (strain Nichols)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Treponemataceae; Treponema; Treponema pallidum; Treponema pallidum subsp. pallidum
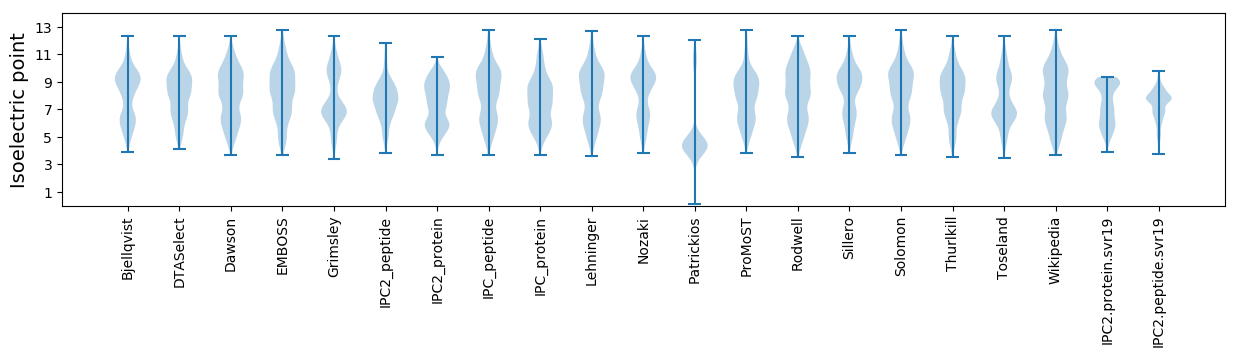
Average proteome isoelectric point is 7.38
Get precalculated fractions of proteins
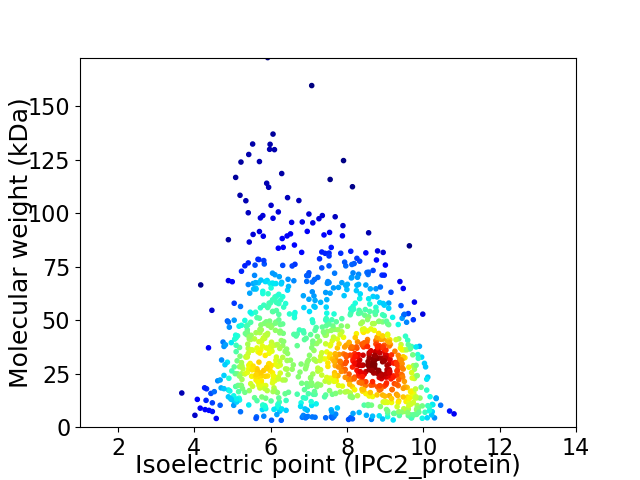
Virtual 2D-PAGE plot for 1027 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O83962|O83962_TREPA Protease IV (SppA) OS=Treponema pallidum (strain Nichols) OX=243276 GN=TP_0997 PE=4 SV=1
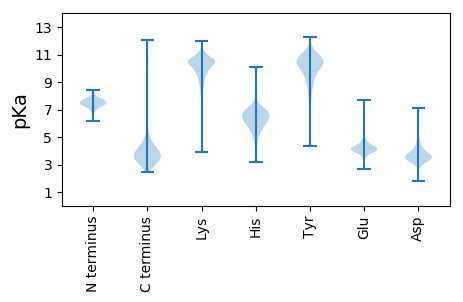
MM1 pKa = 6.89GTYY4 pKa = 8.91MCDD7 pKa = 3.14LCGWGYY13 pKa = 10.59NPEE16 pKa = 4.04VGDD19 pKa = 4.09ADD21 pKa = 3.79GGIPAGTAFEE31 pKa = 4.32NLPDD35 pKa = 3.34HH36 pKa = 6.76WEE38 pKa = 4.33CPLCGVDD45 pKa = 2.96KK46 pKa = 10.93TSFVKK51 pKa = 10.71VV52 pKa = 3.37
MM1 pKa = 6.89GTYY4 pKa = 8.91MCDD7 pKa = 3.14LCGWGYY13 pKa = 10.59NPEE16 pKa = 4.04VGDD19 pKa = 4.09ADD21 pKa = 3.79GGIPAGTAFEE31 pKa = 4.32NLPDD35 pKa = 3.34HH36 pKa = 6.76WEE38 pKa = 4.33CPLCGVDD45 pKa = 2.96KK46 pKa = 10.93TSFVKK51 pKa = 10.71VV52 pKa = 3.37
Molecular weight: 5.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O83367|O83367_TREPA Uncharacterized protein OS=Treponema pallidum (strain Nichols) OX=243276 GN=TP_0347 PE=4 SV=1
MM1 pKa = 7.63SYY3 pKa = 10.43SCSAEE8 pKa = 4.07STSVCSVHH16 pKa = 6.76FSVFRR21 pKa = 11.84IPGATCRR28 pKa = 11.84CDD30 pKa = 3.58VFCAEE35 pKa = 4.78HH36 pKa = 7.07VFFCARR42 pKa = 11.84SQLKK46 pKa = 10.15RR47 pKa = 11.84IVRR50 pKa = 11.84ALCVNGHH57 pKa = 4.91TAKK60 pKa = 10.67FSRR63 pKa = 11.84PLHH66 pKa = 5.11VRR68 pKa = 11.84DD69 pKa = 3.09RR70 pKa = 11.84VSFEE74 pKa = 3.84WVRR77 pKa = 11.84SVPPALIPEE86 pKa = 4.68NISLSILFEE95 pKa = 4.38NEE97 pKa = 4.32DD98 pKa = 3.58IIAVNKK104 pKa = 7.63AQGMIVHH111 pKa = 7.34PGAGHH116 pKa = 4.79WTGTLVQALSFYY128 pKa = 10.25RR129 pKa = 11.84VYY131 pKa = 10.56RR132 pKa = 11.84ARR134 pKa = 11.84FEE136 pKa = 5.42DD137 pKa = 3.8EE138 pKa = 4.73FSRR141 pKa = 11.84QFQKK145 pKa = 11.01GFPDD149 pKa = 4.22FFSTLRR155 pKa = 11.84QGIVHH160 pKa = 7.31RR161 pKa = 11.84LDD163 pKa = 3.82KK164 pKa = 10.49DD165 pKa = 3.56TSGVLLTSRR174 pKa = 11.84NMHH177 pKa = 5.44AHH179 pKa = 5.89EE180 pKa = 4.27ALVRR184 pKa = 11.84SFKK187 pKa = 10.35KK188 pKa = 9.21RR189 pKa = 11.84QVRR192 pKa = 11.84KK193 pKa = 10.21VYY195 pKa = 10.26LALLQGVPARR205 pKa = 11.84GVGVIEE211 pKa = 3.98TTIVRR216 pKa = 11.84DD217 pKa = 3.35RR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84RR221 pKa = 11.84TRR223 pKa = 11.84FVASEE228 pKa = 4.17DD229 pKa = 3.77FSKK232 pKa = 11.1GKK234 pKa = 9.35YY235 pKa = 9.45ARR237 pKa = 11.84TRR239 pKa = 11.84YY240 pKa = 10.07KK241 pKa = 10.76VMKK244 pKa = 10.03ICGACAFVQFLLDD257 pKa = 3.09TGRR260 pKa = 11.84THH262 pKa = 7.41QIRR265 pKa = 11.84VHH267 pKa = 6.06ARR269 pKa = 11.84YY270 pKa = 9.46LGCPVVGDD278 pKa = 3.73PLYY281 pKa = 10.96GSRR284 pKa = 11.84NICGIPTTLMLHH296 pKa = 6.71AYY298 pKa = 9.31AVRR301 pKa = 11.84FVLPRR306 pKa = 11.84TKK308 pKa = 10.59KK309 pKa = 10.45RR310 pKa = 11.84ITLVAPIPLRR320 pKa = 11.84FVRR323 pKa = 11.84LIHH326 pKa = 6.22RR327 pKa = 11.84LSVRR331 pKa = 3.29
MM1 pKa = 7.63SYY3 pKa = 10.43SCSAEE8 pKa = 4.07STSVCSVHH16 pKa = 6.76FSVFRR21 pKa = 11.84IPGATCRR28 pKa = 11.84CDD30 pKa = 3.58VFCAEE35 pKa = 4.78HH36 pKa = 7.07VFFCARR42 pKa = 11.84SQLKK46 pKa = 10.15RR47 pKa = 11.84IVRR50 pKa = 11.84ALCVNGHH57 pKa = 4.91TAKK60 pKa = 10.67FSRR63 pKa = 11.84PLHH66 pKa = 5.11VRR68 pKa = 11.84DD69 pKa = 3.09RR70 pKa = 11.84VSFEE74 pKa = 3.84WVRR77 pKa = 11.84SVPPALIPEE86 pKa = 4.68NISLSILFEE95 pKa = 4.38NEE97 pKa = 4.32DD98 pKa = 3.58IIAVNKK104 pKa = 7.63AQGMIVHH111 pKa = 7.34PGAGHH116 pKa = 4.79WTGTLVQALSFYY128 pKa = 10.25RR129 pKa = 11.84VYY131 pKa = 10.56RR132 pKa = 11.84ARR134 pKa = 11.84FEE136 pKa = 5.42DD137 pKa = 3.8EE138 pKa = 4.73FSRR141 pKa = 11.84QFQKK145 pKa = 11.01GFPDD149 pKa = 4.22FFSTLRR155 pKa = 11.84QGIVHH160 pKa = 7.31RR161 pKa = 11.84LDD163 pKa = 3.82KK164 pKa = 10.49DD165 pKa = 3.56TSGVLLTSRR174 pKa = 11.84NMHH177 pKa = 5.44AHH179 pKa = 5.89EE180 pKa = 4.27ALVRR184 pKa = 11.84SFKK187 pKa = 10.35KK188 pKa = 9.21RR189 pKa = 11.84QVRR192 pKa = 11.84KK193 pKa = 10.21VYY195 pKa = 10.26LALLQGVPARR205 pKa = 11.84GVGVIEE211 pKa = 3.98TTIVRR216 pKa = 11.84DD217 pKa = 3.35RR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84RR221 pKa = 11.84TRR223 pKa = 11.84FVASEE228 pKa = 4.17DD229 pKa = 3.77FSKK232 pKa = 11.1GKK234 pKa = 9.35YY235 pKa = 9.45ARR237 pKa = 11.84TRR239 pKa = 11.84YY240 pKa = 10.07KK241 pKa = 10.76VMKK244 pKa = 10.03ICGACAFVQFLLDD257 pKa = 3.09TGRR260 pKa = 11.84THH262 pKa = 7.41QIRR265 pKa = 11.84VHH267 pKa = 6.06ARR269 pKa = 11.84YY270 pKa = 9.46LGCPVVGDD278 pKa = 3.73PLYY281 pKa = 10.96GSRR284 pKa = 11.84NICGIPTTLMLHH296 pKa = 6.71AYY298 pKa = 9.31AVRR301 pKa = 11.84FVLPRR306 pKa = 11.84TKK308 pKa = 10.59KK309 pKa = 10.45RR310 pKa = 11.84ITLVAPIPLRR320 pKa = 11.84FVRR323 pKa = 11.84LIHH326 pKa = 6.22RR327 pKa = 11.84LSVRR331 pKa = 3.29
Molecular weight: 37.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
349599 |
30 |
1533 |
340.4 |
37.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.124 ± 0.093 | 1.91 ± 0.041 |
4.528 ± 0.05 | 5.981 ± 0.082 |
4.452 ± 0.061 | 6.957 ± 0.073 |
2.757 ± 0.05 | 4.907 ± 0.065 |
3.979 ± 0.064 | 10.152 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.092 ± 0.036 | 2.48 ± 0.036 |
4.203 ± 0.057 | 3.845 ± 0.046 |
7.431 ± 0.077 | 6.622 ± 0.068 |
5.299 ± 0.049 | 8.26 ± 0.081 |
0.977 ± 0.025 | 3.033 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
