
Mosquito VEM virus SDBVL G
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus mosqi1; Mosquito associated gemycircularvirus 1
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
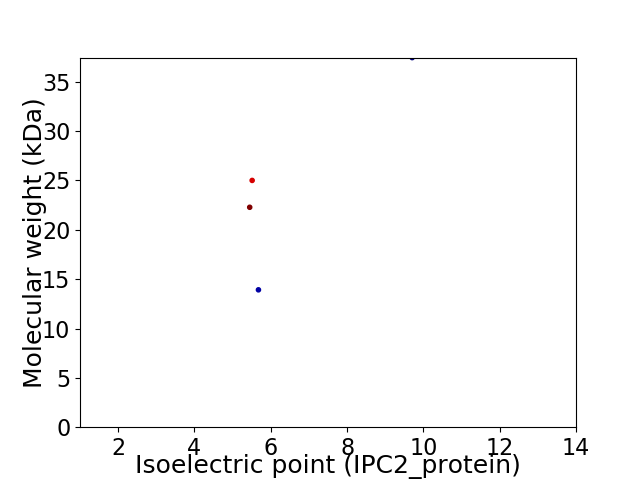
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F6KIE2|F6KIE2_9VIRU Replication-associated protein OS=Mosquito VEM virus SDBVL G OX=1034805 GN=Rep2 PE=4 SV=1
MM1 pKa = 7.63PKK3 pKa = 9.79SLKK6 pKa = 9.86MSLQNCLCDD15 pKa = 3.38SCTSAQNMMPTSSSGRR31 pKa = 11.84NNTPTEE37 pKa = 4.38EE38 pKa = 4.29YY39 pKa = 10.16ISMPFSIFEE48 pKa = 4.65DD49 pKa = 4.0LNSQPEE55 pKa = 4.4TNDD58 pKa = 2.89FGTLPGDD65 pKa = 3.77TPTSQGWDD73 pKa = 2.99ARR75 pKa = 11.84RR76 pKa = 11.84GRR78 pKa = 11.84RR79 pKa = 11.84TITPSRR85 pKa = 11.84TTTSSEE91 pKa = 3.78EE92 pKa = 3.88QPPVLRR98 pKa = 11.84RR99 pKa = 11.84EE100 pKa = 4.07GAALGEE106 pKa = 4.33INNMRR111 pKa = 11.84TGPTSVRR118 pKa = 11.84QFLRR122 pKa = 11.84RR123 pKa = 11.84FSLIEE128 pKa = 4.09SEE130 pKa = 4.55NVDD133 pKa = 3.42PGIWSVLSAILEE145 pKa = 4.0NMQIGTTVLCPSRR158 pKa = 11.84IQLRR162 pKa = 11.84RR163 pKa = 11.84STDD166 pKa = 3.34SHH168 pKa = 5.52WLITLRR174 pKa = 11.84STTGPLPTLEE184 pKa = 4.42EE185 pKa = 3.95MLLPGVYY192 pKa = 9.9HH193 pKa = 6.88PPDD196 pKa = 3.91PRR198 pKa = 11.84QDD200 pKa = 3.07
MM1 pKa = 7.63PKK3 pKa = 9.79SLKK6 pKa = 9.86MSLQNCLCDD15 pKa = 3.38SCTSAQNMMPTSSSGRR31 pKa = 11.84NNTPTEE37 pKa = 4.38EE38 pKa = 4.29YY39 pKa = 10.16ISMPFSIFEE48 pKa = 4.65DD49 pKa = 4.0LNSQPEE55 pKa = 4.4TNDD58 pKa = 2.89FGTLPGDD65 pKa = 3.77TPTSQGWDD73 pKa = 2.99ARR75 pKa = 11.84RR76 pKa = 11.84GRR78 pKa = 11.84RR79 pKa = 11.84TITPSRR85 pKa = 11.84TTTSSEE91 pKa = 3.78EE92 pKa = 3.88QPPVLRR98 pKa = 11.84RR99 pKa = 11.84EE100 pKa = 4.07GAALGEE106 pKa = 4.33INNMRR111 pKa = 11.84TGPTSVRR118 pKa = 11.84QFLRR122 pKa = 11.84RR123 pKa = 11.84FSLIEE128 pKa = 4.09SEE130 pKa = 4.55NVDD133 pKa = 3.42PGIWSVLSAILEE145 pKa = 4.0NMQIGTTVLCPSRR158 pKa = 11.84IQLRR162 pKa = 11.84RR163 pKa = 11.84STDD166 pKa = 3.34SHH168 pKa = 5.52WLITLRR174 pKa = 11.84STTGPLPTLEE184 pKa = 4.42EE185 pKa = 3.95MLLPGVYY192 pKa = 9.9HH193 pKa = 6.88PPDD196 pKa = 3.91PRR198 pKa = 11.84QDD200 pKa = 3.07
Molecular weight: 22.28 kDa
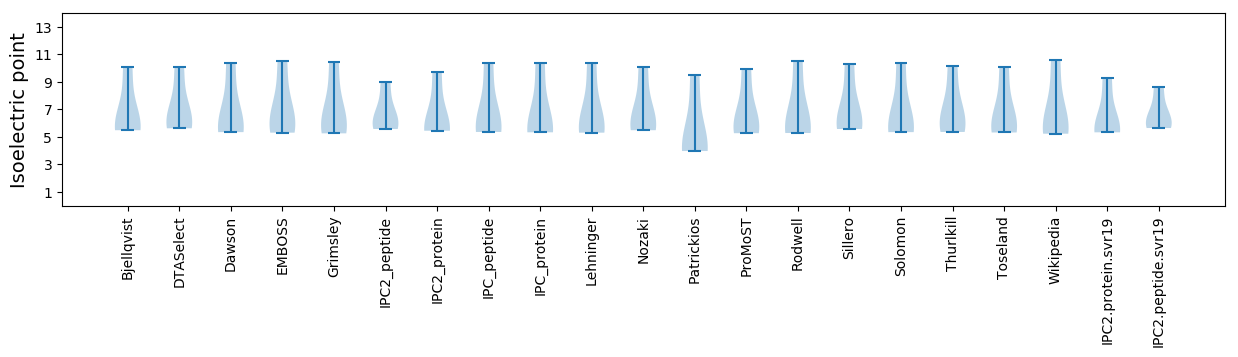
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F6KIE3|F6KIE3_9VIRU Coat protein OS=Mosquito VEM virus SDBVL G OX=1034805 GN=coat protein PE=4 SV=1
MM1 pKa = 7.42VYY3 pKa = 10.0RR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84PTSYY11 pKa = 10.36RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84TGRR17 pKa = 11.84YY18 pKa = 7.63APRR21 pKa = 11.84RR22 pKa = 11.84SIYY25 pKa = 8.8RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 9.75RR29 pKa = 11.84MVYY32 pKa = 9.18RR33 pKa = 11.84RR34 pKa = 11.84ANARR38 pKa = 11.84RR39 pKa = 11.84TRR41 pKa = 11.84TRR43 pKa = 11.84GLPVKK48 pKa = 10.37RR49 pKa = 11.84IRR51 pKa = 11.84NLTSRR56 pKa = 11.84KK57 pKa = 9.74KK58 pKa = 10.15QDD60 pKa = 3.66TMLSWAKK67 pKa = 7.57PTPYY71 pKa = 10.0TSGAVGSPLMTGIPNARR88 pKa = 11.84DD89 pKa = 3.51PGIQPYY95 pKa = 10.5VYY97 pKa = 9.15MYY99 pKa = 10.82QPTARR104 pKa = 11.84DD105 pKa = 3.66LEE107 pKa = 4.35RR108 pKa = 11.84VPGSVTNINDD118 pKa = 3.56SASRR122 pKa = 11.84TSSNCYY128 pKa = 10.08AVGLKK133 pKa = 10.23EE134 pKa = 4.32KK135 pKa = 10.46ISLIVNSNEE144 pKa = 2.97EE145 pKa = 4.24WLWRR149 pKa = 11.84RR150 pKa = 11.84IVFTFKK156 pKa = 9.82GTYY159 pKa = 9.86LVTPQAGFGGDD170 pKa = 3.74SNNVGAFYY178 pKa = 11.03DD179 pKa = 3.88EE180 pKa = 4.54TSVGFPRR187 pKa = 11.84AWTQLSGFTASPGADD202 pKa = 3.17TNALWLALRR211 pKa = 11.84SLLFKK216 pKa = 10.17GTYY219 pKa = 7.1QTDD222 pKa = 3.13WNDD225 pKa = 2.9FATAKK230 pKa = 10.54ADD232 pKa = 3.57NSRR235 pKa = 11.84LNILYY240 pKa = 9.81DD241 pKa = 3.52KK242 pKa = 10.28QVKK245 pKa = 8.59VQSPNDD251 pKa = 3.38NGTTRR256 pKa = 11.84TFNRR260 pKa = 11.84WHH262 pKa = 7.36PFRR265 pKa = 11.84KK266 pKa = 9.59RR267 pKa = 11.84LIYY270 pKa = 10.76NDD272 pKa = 4.2DD273 pKa = 3.53EE274 pKa = 4.88VGGGKK279 pKa = 9.48QVDD282 pKa = 3.53VGYY285 pKa = 11.0SSMHH289 pKa = 6.09RR290 pKa = 11.84ASMGDD295 pKa = 3.03AYY297 pKa = 11.02VFDD300 pKa = 4.3MFIPGHH306 pKa = 5.83NSEE309 pKa = 4.42QADD312 pKa = 4.0EE313 pKa = 4.87LEE315 pKa = 4.65VRR317 pKa = 11.84CSSTYY322 pKa = 9.76YY323 pKa = 8.97WHH325 pKa = 6.35EE326 pKa = 3.98TT327 pKa = 3.35
MM1 pKa = 7.42VYY3 pKa = 10.0RR4 pKa = 11.84RR5 pKa = 11.84SRR7 pKa = 11.84PTSYY11 pKa = 10.36RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84TGRR17 pKa = 11.84YY18 pKa = 7.63APRR21 pKa = 11.84RR22 pKa = 11.84SIYY25 pKa = 8.8RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 9.75RR29 pKa = 11.84MVYY32 pKa = 9.18RR33 pKa = 11.84RR34 pKa = 11.84ANARR38 pKa = 11.84RR39 pKa = 11.84TRR41 pKa = 11.84TRR43 pKa = 11.84GLPVKK48 pKa = 10.37RR49 pKa = 11.84IRR51 pKa = 11.84NLTSRR56 pKa = 11.84KK57 pKa = 9.74KK58 pKa = 10.15QDD60 pKa = 3.66TMLSWAKK67 pKa = 7.57PTPYY71 pKa = 10.0TSGAVGSPLMTGIPNARR88 pKa = 11.84DD89 pKa = 3.51PGIQPYY95 pKa = 10.5VYY97 pKa = 9.15MYY99 pKa = 10.82QPTARR104 pKa = 11.84DD105 pKa = 3.66LEE107 pKa = 4.35RR108 pKa = 11.84VPGSVTNINDD118 pKa = 3.56SASRR122 pKa = 11.84TSSNCYY128 pKa = 10.08AVGLKK133 pKa = 10.23EE134 pKa = 4.32KK135 pKa = 10.46ISLIVNSNEE144 pKa = 2.97EE145 pKa = 4.24WLWRR149 pKa = 11.84RR150 pKa = 11.84IVFTFKK156 pKa = 9.82GTYY159 pKa = 9.86LVTPQAGFGGDD170 pKa = 3.74SNNVGAFYY178 pKa = 11.03DD179 pKa = 3.88EE180 pKa = 4.54TSVGFPRR187 pKa = 11.84AWTQLSGFTASPGADD202 pKa = 3.17TNALWLALRR211 pKa = 11.84SLLFKK216 pKa = 10.17GTYY219 pKa = 7.1QTDD222 pKa = 3.13WNDD225 pKa = 2.9FATAKK230 pKa = 10.54ADD232 pKa = 3.57NSRR235 pKa = 11.84LNILYY240 pKa = 9.81DD241 pKa = 3.52KK242 pKa = 10.28QVKK245 pKa = 8.59VQSPNDD251 pKa = 3.38NGTTRR256 pKa = 11.84TFNRR260 pKa = 11.84WHH262 pKa = 7.36PFRR265 pKa = 11.84KK266 pKa = 9.59RR267 pKa = 11.84LIYY270 pKa = 10.76NDD272 pKa = 4.2DD273 pKa = 3.53EE274 pKa = 4.88VGGGKK279 pKa = 9.48QVDD282 pKa = 3.53VGYY285 pKa = 11.0SSMHH289 pKa = 6.09RR290 pKa = 11.84ASMGDD295 pKa = 3.03AYY297 pKa = 11.02VFDD300 pKa = 4.3MFIPGHH306 pKa = 5.83NSEE309 pKa = 4.42QADD312 pKa = 4.0EE313 pKa = 4.87LEE315 pKa = 4.65VRR317 pKa = 11.84CSSTYY322 pKa = 9.76YY323 pKa = 8.97WHH325 pKa = 6.35EE326 pKa = 3.98TT327 pKa = 3.35
Molecular weight: 37.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
869 |
121 |
327 |
217.3 |
24.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.869 ± 1.033 | 1.496 ± 0.417 |
5.869 ± 0.589 | 5.409 ± 1.075 |
4.373 ± 0.702 | 7.48 ± 0.546 |
1.956 ± 0.547 | 4.373 ± 0.583 |
3.797 ± 0.84 | 6.904 ± 0.943 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.301 ± 0.64 | 4.488 ± 0.676 |
6.559 ± 0.907 | 2.877 ± 0.553 |
8.746 ± 1.265 | 8.055 ± 1.067 |
7.48 ± 1.141 | 5.524 ± 0.732 |
2.417 ± 0.326 | 4.028 ± 1.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
