
Hyphomonas jannaschiana VP2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomonadales; Hyphomonadaceae; Hyphomonas; Hyphomonas jannaschiana
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
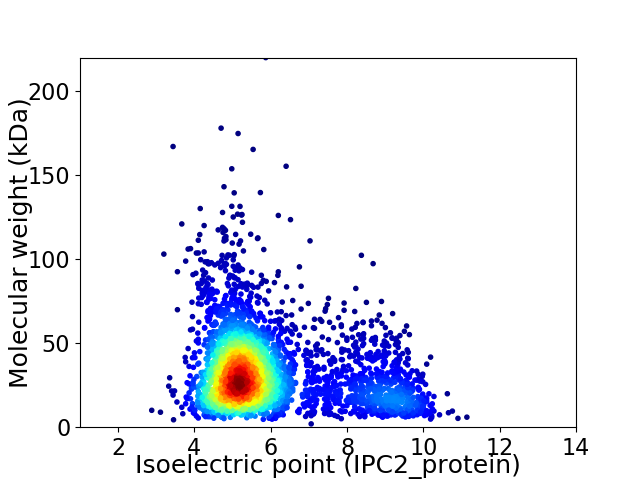
Virtual 2D-PAGE plot for 3505 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A059FJU2|A0A059FJU2_9PROT Tryptophanase/L-cysteine desulfhydrase PLP-dependent OS=Hyphomonas jannaschiana VP2 OX=1280952 GN=tnaA PE=3 SV=1
MM1 pKa = 7.87KK2 pKa = 10.42SRR4 pKa = 11.84LLASSVFAGAAFMAQASMLAVAQEE28 pKa = 3.78ADD30 pKa = 3.61EE31 pKa = 5.21AVDD34 pKa = 3.98TVATTTEE41 pKa = 4.14DD42 pKa = 2.82ATARR46 pKa = 11.84QEE48 pKa = 4.25TVYY51 pKa = 9.58VTGSRR56 pKa = 11.84IAQPNMTTTSPVTSVSANDD75 pKa = 3.36VKK77 pKa = 11.11LQGVTRR83 pKa = 11.84VEE85 pKa = 4.23DD86 pKa = 4.08LVTQLPQAFAAQNSTVSNGASGTATVSLRR115 pKa = 11.84NLDD118 pKa = 3.81LGGSATRR125 pKa = 11.84TLVLVDD131 pKa = 4.2GKK133 pKa = 10.23RR134 pKa = 11.84LPYY137 pKa = 10.38GSPNDD142 pKa = 3.65AAADD146 pKa = 3.88LNLIPGQMVEE156 pKa = 4.32RR157 pKa = 11.84VDD159 pKa = 3.84VLTGGASAVYY169 pKa = 10.16GSDD172 pKa = 3.67AIAGVVNFIMKK183 pKa = 10.29DD184 pKa = 3.28DD185 pKa = 4.09FEE187 pKa = 4.75GVQIDD192 pKa = 3.96TQYY195 pKa = 11.88GFYY198 pKa = 10.19QHH200 pKa = 6.61NNDD203 pKa = 3.43YY204 pKa = 8.78DD205 pKa = 4.13TNGDD209 pKa = 3.67LRR211 pKa = 11.84DD212 pKa = 3.99VIAQRR217 pKa = 11.84AVGNPSQFALPDD229 pKa = 4.04DD230 pKa = 4.17NVIDD234 pKa = 4.89GYY236 pKa = 10.9SKK238 pKa = 10.84EE239 pKa = 3.69ITAIMGVNTDD249 pKa = 4.01DD250 pKa = 4.37GRR252 pKa = 11.84GNLTAWFGYY261 pKa = 9.85RR262 pKa = 11.84NNDD265 pKa = 2.84EE266 pKa = 4.38VLQGDD271 pKa = 4.18RR272 pKa = 11.84DD273 pKa = 3.87YY274 pKa = 11.25SACAIGSATATGFTCGGSSTSFPGRR299 pKa = 11.84FTDD302 pKa = 5.61FDD304 pKa = 3.99AFNYY308 pKa = 9.8TIDD311 pKa = 3.79PATGEE316 pKa = 4.08FRR318 pKa = 11.84DD319 pKa = 4.16FDD321 pKa = 4.22AATDD325 pKa = 3.65QYY327 pKa = 11.78NYY329 pKa = 10.94GPLNFYY335 pKa = 10.2QRR337 pKa = 11.84PDD339 pKa = 3.08EE340 pKa = 4.62RR341 pKa = 11.84YY342 pKa = 6.83TAGFNGHH349 pKa = 5.44YY350 pKa = 9.98QINDD354 pKa = 3.14YY355 pKa = 11.44AEE357 pKa = 5.38AYY359 pKa = 6.49TQFMFMDD366 pKa = 4.12YY367 pKa = 10.78QSNAQIAPSGNFFATTSINCDD388 pKa = 3.12NPLISASQLADD399 pKa = 3.24IGCTSANIAAGDD411 pKa = 4.13SIGMYY416 pKa = 8.89IARR419 pKa = 11.84RR420 pKa = 11.84NVEE423 pKa = 3.56GGGRR427 pKa = 11.84QDD429 pKa = 3.18HH430 pKa = 6.83LGYY433 pKa = 8.35QTYY436 pKa = 10.45RR437 pKa = 11.84GVAGLRR443 pKa = 11.84GDD445 pKa = 4.48LYY447 pKa = 11.05KK448 pKa = 11.06APGWSYY454 pKa = 11.17DD455 pKa = 2.93ISASFAQVTLSRR467 pKa = 11.84SYY469 pKa = 11.12RR470 pKa = 11.84NEE472 pKa = 3.49FSITRR477 pKa = 11.84LNRR480 pKa = 11.84ALNVVDD486 pKa = 5.59DD487 pKa = 4.38GTGTAVCASVLDD499 pKa = 4.26GTDD502 pKa = 3.78PNCVPWDD509 pKa = 3.27IFTVGNVTPEE519 pKa = 3.68ALAYY523 pKa = 10.3LQVPLLQTGTTEE535 pKa = 3.88QNIVSGVLTGDD546 pKa = 3.79LGQYY550 pKa = 9.63GFQLPSANSGLQVAIGAEE568 pKa = 3.98YY569 pKa = 10.47RR570 pKa = 11.84RR571 pKa = 11.84DD572 pKa = 3.67ALEE575 pKa = 4.78SITDD579 pKa = 3.5NSFATGDD586 pKa = 3.66GAGQGGPTIGLSGAVDD602 pKa = 3.89SYY604 pKa = 12.28DD605 pKa = 3.33MFAEE609 pKa = 4.3FQMPLVEE616 pKa = 4.48GKK618 pKa = 9.74PGVEE622 pKa = 3.89LLSIEE627 pKa = 4.03GAYY630 pKa = 9.94RR631 pKa = 11.84YY632 pKa = 10.02SDD634 pKa = 3.71YY635 pKa = 10.94STGISADD642 pKa = 3.43SYY644 pKa = 11.55KK645 pKa = 10.48IGGDD649 pKa = 3.61YY650 pKa = 11.13APTSDD655 pKa = 2.57IRR657 pKa = 11.84FRR659 pKa = 11.84ASFQKK664 pKa = 10.53AVRR667 pKa = 11.84APNVIDD673 pKa = 4.84LFQAQGFNLFDD684 pKa = 6.12LDD686 pKa = 6.14DD687 pKa = 4.94DD688 pKa = 4.9LCDD691 pKa = 3.95FTDD694 pKa = 3.93PAGDD698 pKa = 3.8GTGGAACIGTNPWQVTQAQADD719 pKa = 4.03GGALGSPAGQYY730 pKa = 10.94NYY732 pKa = 10.71LQGGNPDD739 pKa = 3.89LQPEE743 pKa = 4.16EE744 pKa = 5.61AEE746 pKa = 4.09TLTIGFVATPTFLPGFSASVDD767 pKa = 3.67YY768 pKa = 11.36YY769 pKa = 11.49NIDD772 pKa = 3.11ITDD775 pKa = 5.07AISTVGSSVTMQLCYY790 pKa = 10.58LQGDD794 pKa = 3.74ADD796 pKa = 3.77ACQRR800 pKa = 11.84IQRR803 pKa = 11.84NANGQLWVGTGNVVDD818 pKa = 4.84LNVNIGGVEE827 pKa = 3.85TSGFDD832 pKa = 2.89ISAAYY837 pKa = 9.45GFDD840 pKa = 3.07MGKK843 pKa = 9.59YY844 pKa = 9.96GSMNLSMNGTLLDD857 pKa = 4.14SFDD860 pKa = 3.76VDD862 pKa = 3.8PVGNAAYY869 pKa = 9.8AYY871 pKa = 10.02DD872 pKa = 3.89CVGKK876 pKa = 10.77YY877 pKa = 10.94GNDD880 pKa = 3.47CFTPTPEE887 pKa = 3.44WRR889 pKa = 11.84HH890 pKa = 5.2RR891 pKa = 11.84ARR893 pKa = 11.84ASWVTPVDD901 pKa = 3.74GLDD904 pKa = 4.66LSATWRR910 pKa = 11.84YY911 pKa = 9.52IGSADD916 pKa = 4.69LDD918 pKa = 3.67TGVTNRR924 pKa = 11.84VDD926 pKa = 3.52STLEE930 pKa = 3.65EE931 pKa = 3.81QNYY934 pKa = 9.8IDD936 pKa = 6.41LAGQWAAKK944 pKa = 9.84DD945 pKa = 3.36WVSFRR950 pKa = 11.84FGVNNVLDD958 pKa = 4.89DD959 pKa = 5.01DD960 pKa = 5.08PPLSASTGTTGNGNTYY976 pKa = 8.89PQTYY980 pKa = 9.88DD981 pKa = 2.69AMGRR985 pKa = 11.84YY986 pKa = 8.69FFVGATFDD994 pKa = 3.57FF995 pKa = 4.93
MM1 pKa = 7.87KK2 pKa = 10.42SRR4 pKa = 11.84LLASSVFAGAAFMAQASMLAVAQEE28 pKa = 3.78ADD30 pKa = 3.61EE31 pKa = 5.21AVDD34 pKa = 3.98TVATTTEE41 pKa = 4.14DD42 pKa = 2.82ATARR46 pKa = 11.84QEE48 pKa = 4.25TVYY51 pKa = 9.58VTGSRR56 pKa = 11.84IAQPNMTTTSPVTSVSANDD75 pKa = 3.36VKK77 pKa = 11.11LQGVTRR83 pKa = 11.84VEE85 pKa = 4.23DD86 pKa = 4.08LVTQLPQAFAAQNSTVSNGASGTATVSLRR115 pKa = 11.84NLDD118 pKa = 3.81LGGSATRR125 pKa = 11.84TLVLVDD131 pKa = 4.2GKK133 pKa = 10.23RR134 pKa = 11.84LPYY137 pKa = 10.38GSPNDD142 pKa = 3.65AAADD146 pKa = 3.88LNLIPGQMVEE156 pKa = 4.32RR157 pKa = 11.84VDD159 pKa = 3.84VLTGGASAVYY169 pKa = 10.16GSDD172 pKa = 3.67AIAGVVNFIMKK183 pKa = 10.29DD184 pKa = 3.28DD185 pKa = 4.09FEE187 pKa = 4.75GVQIDD192 pKa = 3.96TQYY195 pKa = 11.88GFYY198 pKa = 10.19QHH200 pKa = 6.61NNDD203 pKa = 3.43YY204 pKa = 8.78DD205 pKa = 4.13TNGDD209 pKa = 3.67LRR211 pKa = 11.84DD212 pKa = 3.99VIAQRR217 pKa = 11.84AVGNPSQFALPDD229 pKa = 4.04DD230 pKa = 4.17NVIDD234 pKa = 4.89GYY236 pKa = 10.9SKK238 pKa = 10.84EE239 pKa = 3.69ITAIMGVNTDD249 pKa = 4.01DD250 pKa = 4.37GRR252 pKa = 11.84GNLTAWFGYY261 pKa = 9.85RR262 pKa = 11.84NNDD265 pKa = 2.84EE266 pKa = 4.38VLQGDD271 pKa = 4.18RR272 pKa = 11.84DD273 pKa = 3.87YY274 pKa = 11.25SACAIGSATATGFTCGGSSTSFPGRR299 pKa = 11.84FTDD302 pKa = 5.61FDD304 pKa = 3.99AFNYY308 pKa = 9.8TIDD311 pKa = 3.79PATGEE316 pKa = 4.08FRR318 pKa = 11.84DD319 pKa = 4.16FDD321 pKa = 4.22AATDD325 pKa = 3.65QYY327 pKa = 11.78NYY329 pKa = 10.94GPLNFYY335 pKa = 10.2QRR337 pKa = 11.84PDD339 pKa = 3.08EE340 pKa = 4.62RR341 pKa = 11.84YY342 pKa = 6.83TAGFNGHH349 pKa = 5.44YY350 pKa = 9.98QINDD354 pKa = 3.14YY355 pKa = 11.44AEE357 pKa = 5.38AYY359 pKa = 6.49TQFMFMDD366 pKa = 4.12YY367 pKa = 10.78QSNAQIAPSGNFFATTSINCDD388 pKa = 3.12NPLISASQLADD399 pKa = 3.24IGCTSANIAAGDD411 pKa = 4.13SIGMYY416 pKa = 8.89IARR419 pKa = 11.84RR420 pKa = 11.84NVEE423 pKa = 3.56GGGRR427 pKa = 11.84QDD429 pKa = 3.18HH430 pKa = 6.83LGYY433 pKa = 8.35QTYY436 pKa = 10.45RR437 pKa = 11.84GVAGLRR443 pKa = 11.84GDD445 pKa = 4.48LYY447 pKa = 11.05KK448 pKa = 11.06APGWSYY454 pKa = 11.17DD455 pKa = 2.93ISASFAQVTLSRR467 pKa = 11.84SYY469 pKa = 11.12RR470 pKa = 11.84NEE472 pKa = 3.49FSITRR477 pKa = 11.84LNRR480 pKa = 11.84ALNVVDD486 pKa = 5.59DD487 pKa = 4.38GTGTAVCASVLDD499 pKa = 4.26GTDD502 pKa = 3.78PNCVPWDD509 pKa = 3.27IFTVGNVTPEE519 pKa = 3.68ALAYY523 pKa = 10.3LQVPLLQTGTTEE535 pKa = 3.88QNIVSGVLTGDD546 pKa = 3.79LGQYY550 pKa = 9.63GFQLPSANSGLQVAIGAEE568 pKa = 3.98YY569 pKa = 10.47RR570 pKa = 11.84RR571 pKa = 11.84DD572 pKa = 3.67ALEE575 pKa = 4.78SITDD579 pKa = 3.5NSFATGDD586 pKa = 3.66GAGQGGPTIGLSGAVDD602 pKa = 3.89SYY604 pKa = 12.28DD605 pKa = 3.33MFAEE609 pKa = 4.3FQMPLVEE616 pKa = 4.48GKK618 pKa = 9.74PGVEE622 pKa = 3.89LLSIEE627 pKa = 4.03GAYY630 pKa = 9.94RR631 pKa = 11.84YY632 pKa = 10.02SDD634 pKa = 3.71YY635 pKa = 10.94STGISADD642 pKa = 3.43SYY644 pKa = 11.55KK645 pKa = 10.48IGGDD649 pKa = 3.61YY650 pKa = 11.13APTSDD655 pKa = 2.57IRR657 pKa = 11.84FRR659 pKa = 11.84ASFQKK664 pKa = 10.53AVRR667 pKa = 11.84APNVIDD673 pKa = 4.84LFQAQGFNLFDD684 pKa = 6.12LDD686 pKa = 6.14DD687 pKa = 4.94DD688 pKa = 4.9LCDD691 pKa = 3.95FTDD694 pKa = 3.93PAGDD698 pKa = 3.8GTGGAACIGTNPWQVTQAQADD719 pKa = 4.03GGALGSPAGQYY730 pKa = 10.94NYY732 pKa = 10.71LQGGNPDD739 pKa = 3.89LQPEE743 pKa = 4.16EE744 pKa = 5.61AEE746 pKa = 4.09TLTIGFVATPTFLPGFSASVDD767 pKa = 3.67YY768 pKa = 11.36YY769 pKa = 11.49NIDD772 pKa = 3.11ITDD775 pKa = 5.07AISTVGSSVTMQLCYY790 pKa = 10.58LQGDD794 pKa = 3.74ADD796 pKa = 3.77ACQRR800 pKa = 11.84IQRR803 pKa = 11.84NANGQLWVGTGNVVDD818 pKa = 4.84LNVNIGGVEE827 pKa = 3.85TSGFDD832 pKa = 2.89ISAAYY837 pKa = 9.45GFDD840 pKa = 3.07MGKK843 pKa = 9.59YY844 pKa = 9.96GSMNLSMNGTLLDD857 pKa = 4.14SFDD860 pKa = 3.76VDD862 pKa = 3.8PVGNAAYY869 pKa = 9.8AYY871 pKa = 10.02DD872 pKa = 3.89CVGKK876 pKa = 10.77YY877 pKa = 10.94GNDD880 pKa = 3.47CFTPTPEE887 pKa = 3.44WRR889 pKa = 11.84HH890 pKa = 5.2RR891 pKa = 11.84ARR893 pKa = 11.84ASWVTPVDD901 pKa = 3.74GLDD904 pKa = 4.66LSATWRR910 pKa = 11.84YY911 pKa = 9.52IGSADD916 pKa = 4.69LDD918 pKa = 3.67TGVTNRR924 pKa = 11.84VDD926 pKa = 3.52STLEE930 pKa = 3.65EE931 pKa = 3.81QNYY934 pKa = 9.8IDD936 pKa = 6.41LAGQWAAKK944 pKa = 9.84DD945 pKa = 3.36WVSFRR950 pKa = 11.84FGVNNVLDD958 pKa = 4.89DD959 pKa = 5.01DD960 pKa = 5.08PPLSASTGTTGNGNTYY976 pKa = 8.89PQTYY980 pKa = 9.88DD981 pKa = 2.69AMGRR985 pKa = 11.84YY986 pKa = 8.69FFVGATFDD994 pKa = 3.57FF995 pKa = 4.93
Molecular weight: 106.13 kDa
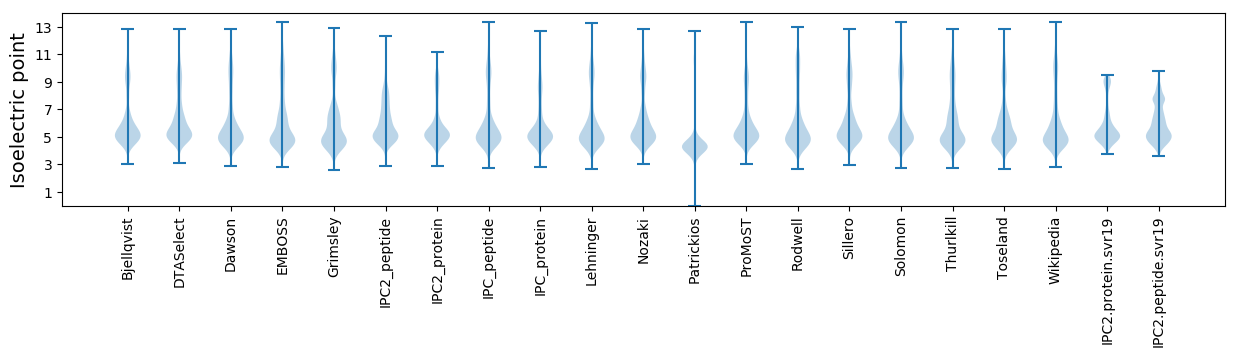
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A059FKR9|A0A059FKR9_9PROT Penicillin-binding protein 2 OS=Hyphomonas jannaschiana VP2 OX=1280952 GN=HJA_00830 PE=4 SV=1
MM1 pKa = 7.44SKK3 pKa = 10.4SPNSVLHH10 pKa = 5.94PAKK13 pKa = 10.23KK14 pKa = 9.76PRR16 pKa = 11.84VKK18 pKa = 10.33RR19 pKa = 11.84RR20 pKa = 11.84NATWKK25 pKa = 10.6ANLGKK30 pKa = 9.88RR31 pKa = 11.84AKK33 pKa = 10.47VRR35 pKa = 11.84VFGNTKK41 pKa = 9.19RR42 pKa = 11.84ARR44 pKa = 11.84IARR47 pKa = 11.84KK48 pKa = 9.21KK49 pKa = 10.51AAGKK53 pKa = 10.03
MM1 pKa = 7.44SKK3 pKa = 10.4SPNSVLHH10 pKa = 5.94PAKK13 pKa = 10.23KK14 pKa = 9.76PRR16 pKa = 11.84VKK18 pKa = 10.33RR19 pKa = 11.84RR20 pKa = 11.84NATWKK25 pKa = 10.6ANLGKK30 pKa = 9.88RR31 pKa = 11.84AKK33 pKa = 10.47VRR35 pKa = 11.84VFGNTKK41 pKa = 9.19RR42 pKa = 11.84ARR44 pKa = 11.84IARR47 pKa = 11.84KK48 pKa = 9.21KK49 pKa = 10.51AAGKK53 pKa = 10.03
Molecular weight: 5.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1100037 |
19 |
2026 |
313.8 |
34.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.059 ± 0.056 | 0.843 ± 0.012 |
6.206 ± 0.039 | 6.35 ± 0.039 |
3.826 ± 0.026 | 8.595 ± 0.041 |
1.908 ± 0.019 | 5.091 ± 0.031 |
3.634 ± 0.039 | 9.673 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.659 ± 0.021 | 2.712 ± 0.022 |
5.191 ± 0.031 | 3.08 ± 0.019 |
6.407 ± 0.041 | 5.534 ± 0.03 |
5.4 ± 0.031 | 6.999 ± 0.031 |
1.419 ± 0.018 | 2.416 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
