
Wenzhou yanvirus-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.54
Get precalculated fractions of proteins
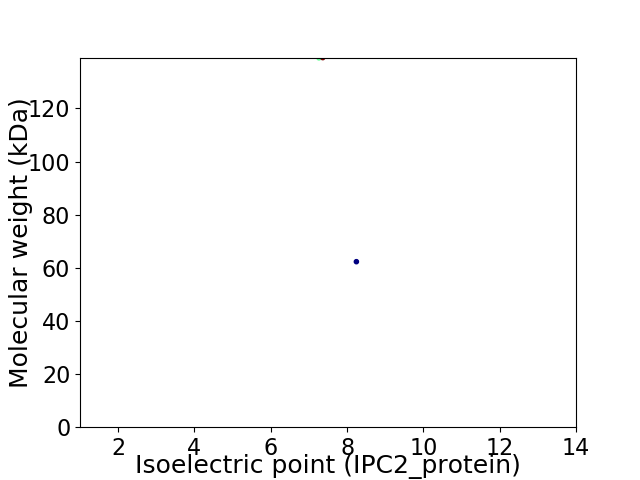
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLD2|A0A1L3KLD2_9VIRU Uncharacterized protein OS=Wenzhou yanvirus-like virus 2 OX=1923684 PE=4 SV=1
MM1 pKa = 7.92SITTHH6 pKa = 6.06KK7 pKa = 9.61KK8 pKa = 9.53KK9 pKa = 10.56CGSHH13 pKa = 6.81PDD15 pKa = 3.41VKK17 pKa = 10.64RR18 pKa = 11.84GAEE21 pKa = 3.5QVLFQKK27 pKa = 10.71YY28 pKa = 7.75HH29 pKa = 4.84GKK31 pKa = 9.9YY32 pKa = 8.36GRR34 pKa = 11.84SRR36 pKa = 11.84RR37 pKa = 11.84SYY39 pKa = 10.02LKK41 pKa = 10.54RR42 pKa = 11.84GFGPDD47 pKa = 3.42GGPDD51 pKa = 2.76RR52 pKa = 11.84AVALVHH58 pKa = 6.71RR59 pKa = 11.84LYY61 pKa = 11.09AGLRR65 pKa = 11.84GAGRR69 pKa = 11.84YY70 pKa = 7.52PVPQNPSDD78 pKa = 3.71ILNHH82 pKa = 6.4LLATNVIDD90 pKa = 3.84GRR92 pKa = 11.84YY93 pKa = 9.02RR94 pKa = 11.84DD95 pKa = 3.92EE96 pKa = 4.36EE97 pKa = 4.22KK98 pKa = 11.05GGLGLPCSLTAFATAFPNQRR118 pKa = 11.84HH119 pKa = 5.35ICLFYY124 pKa = 11.14EE125 pKa = 4.46KK126 pKa = 10.51LCEE129 pKa = 4.22QGLHH133 pKa = 6.04EE134 pKa = 4.89EE135 pKa = 4.3NSRR138 pKa = 11.84GPSGGMTLTGMVSALQEE155 pKa = 3.89WEE157 pKa = 3.94AGIIADD163 pKa = 4.52PKK165 pKa = 9.39PKK167 pKa = 9.98VVRR170 pKa = 11.84PRR172 pKa = 11.84GMDD175 pKa = 3.32LHH177 pKa = 6.48QVNCRR182 pKa = 11.84GGTFLSYY189 pKa = 11.03DD190 pKa = 3.62VVHH193 pKa = 6.19RR194 pKa = 11.84TLLRR198 pKa = 11.84DD199 pKa = 3.49GARR202 pKa = 11.84EE203 pKa = 3.79QIIVYY208 pKa = 10.39INDD211 pKa = 3.16EE212 pKa = 4.3SCNFGHH218 pKa = 6.62FCAVSSLNEE227 pKa = 3.76DD228 pKa = 2.95AMKK231 pKa = 10.81RR232 pKa = 11.84EE233 pKa = 3.66IAKK236 pKa = 9.79GKK238 pKa = 10.06RR239 pKa = 11.84RR240 pKa = 11.84AEE242 pKa = 4.11YY243 pKa = 10.89ADD245 pKa = 3.64MVAADD250 pKa = 3.95SDD252 pKa = 4.09QVPKK256 pKa = 10.6EE257 pKa = 4.11VAPKK261 pKa = 10.23AISGDD266 pKa = 3.92HH267 pKa = 6.14IVPATTSEE275 pKa = 4.27PGPSGINQEE284 pKa = 4.48AEE286 pKa = 4.08THH288 pKa = 6.11HH289 pKa = 6.23SQEE292 pKa = 3.72LDD294 pKa = 3.26ALLEE298 pKa = 3.92EE299 pKa = 5.93AIAEE303 pKa = 4.25MQGMDD308 pKa = 4.51PPQPPPPEE316 pKa = 4.82PNDD319 pKa = 3.4NGPGGNIGGEE329 pKa = 4.03GDD331 pKa = 4.52GPDD334 pKa = 3.69EE335 pKa = 4.14PPIIKK340 pKa = 10.34EE341 pKa = 4.14EE342 pKa = 4.24VPPPPPPPPPEE353 pKa = 4.25PKK355 pKa = 9.75PIPVSRR361 pKa = 11.84RR362 pKa = 11.84TLRR365 pKa = 11.84YY366 pKa = 9.38LRR368 pKa = 11.84EE369 pKa = 3.67KK370 pKa = 11.23LEE372 pKa = 4.46FIKK375 pKa = 9.33THH377 pKa = 5.83SATPILDD384 pKa = 3.56HH385 pKa = 6.82RR386 pKa = 11.84PVRR389 pKa = 11.84AYY391 pKa = 10.32ACAGLVSEE399 pKa = 5.09GCVEE403 pKa = 4.49HH404 pKa = 7.39EE405 pKa = 4.51DD406 pKa = 3.78ASHH409 pKa = 6.4EE410 pKa = 4.34GGVGLTEE417 pKa = 3.97LHH419 pKa = 6.75PEE421 pKa = 3.65VLARR425 pKa = 11.84FNFTRR430 pKa = 11.84RR431 pKa = 11.84SYY433 pKa = 10.35YY434 pKa = 9.78YY435 pKa = 8.23VTAGVEE441 pKa = 3.92PDD443 pKa = 3.47ALDD446 pKa = 4.56GRR448 pKa = 11.84FLTNGQYY455 pKa = 10.56NPAWVASIQFEE466 pKa = 4.31DD467 pKa = 3.75TAFTLEE473 pKa = 4.01FVGNKK478 pKa = 9.61GPCDD482 pKa = 3.5YY483 pKa = 11.19YY484 pKa = 11.15RR485 pKa = 11.84LYY487 pKa = 10.83KK488 pKa = 10.57AATLGAKK495 pKa = 9.79KK496 pKa = 10.64GFFSCPDD503 pKa = 4.68FINEE507 pKa = 4.18ALDD510 pKa = 5.4DD511 pKa = 4.09VSAWAAEE518 pKa = 4.12IGLITQPQATIKK530 pKa = 10.59HH531 pKa = 4.66VTNLPGRR538 pKa = 11.84GLRR541 pKa = 11.84PRR543 pKa = 11.84SLKK546 pKa = 10.46GVAGASRR553 pKa = 11.84AQWAMHH559 pKa = 6.24ARR561 pKa = 11.84HH562 pKa = 6.25APEE565 pKa = 4.66DD566 pKa = 3.51LRR568 pKa = 11.84GPYY571 pKa = 9.82HH572 pKa = 7.15SLFIEE577 pKa = 4.37NADD580 pKa = 4.96DD581 pKa = 4.69SNSDD585 pKa = 3.55PNEE588 pKa = 3.57VLKK591 pKa = 10.88ALKK594 pKa = 10.1YY595 pKa = 9.47EE596 pKa = 3.87KK597 pKa = 11.29AMMEE601 pKa = 4.15KK602 pKa = 9.68EE603 pKa = 3.87HH604 pKa = 6.64GKK606 pKa = 10.2RR607 pKa = 11.84GYY609 pKa = 9.57CAPKK613 pKa = 9.08MSTPKK618 pKa = 9.27TCASCGVSPPKK629 pKa = 10.57GKK631 pKa = 10.71YY632 pKa = 8.04KK633 pKa = 9.94WKK635 pKa = 10.1HH636 pKa = 4.92RR637 pKa = 11.84QCKK640 pKa = 10.1DD641 pKa = 3.2CQQKK645 pKa = 10.8LKK647 pKa = 9.71TCGAVSTMGQQIQQNLTVAEE667 pKa = 4.07GHH669 pKa = 6.83PGCVHH674 pKa = 6.74LNSSTLPPKK683 pKa = 9.87KK684 pKa = 10.15EE685 pKa = 3.47KK686 pKa = 8.47WAKK689 pKa = 8.66VHH691 pKa = 6.11IPKK694 pKa = 10.47GAIKK698 pKa = 9.75VHH700 pKa = 6.44KK701 pKa = 10.43SDD703 pKa = 4.53VPWLRR708 pKa = 11.84HH709 pKa = 5.65ADD711 pKa = 3.12WGGSKK716 pKa = 8.22MHH718 pKa = 6.24SVEE721 pKa = 5.46KK722 pKa = 10.55EE723 pKa = 3.92DD724 pKa = 4.56LSKK727 pKa = 10.54IDD729 pKa = 3.61TTLEE733 pKa = 3.81RR734 pKa = 11.84PKK736 pKa = 10.46RR737 pKa = 11.84EE738 pKa = 3.98CVLAGIGVSGCYY750 pKa = 10.11PMVTRR755 pKa = 11.84KK756 pKa = 10.05GFYY759 pKa = 10.85ARR761 pKa = 11.84MQALIGRR768 pKa = 11.84AYY770 pKa = 9.78LQKK773 pKa = 10.52PEE775 pKa = 4.29SSPAAWEE782 pKa = 4.48AMNQMKK788 pKa = 10.47DD789 pKa = 3.36CLLPNDD795 pKa = 4.64ALDD798 pKa = 4.09GEE800 pKa = 4.68KK801 pKa = 10.7FSIDD805 pKa = 3.12FWLTTMPARR814 pKa = 11.84RR815 pKa = 11.84RR816 pKa = 11.84RR817 pKa = 11.84ALEE820 pKa = 4.04RR821 pKa = 11.84AHH823 pKa = 7.26KK824 pKa = 9.64EE825 pKa = 3.97YY826 pKa = 10.76KK827 pKa = 10.27EE828 pKa = 4.16SGGLRR833 pKa = 11.84DD834 pKa = 3.79KK835 pKa = 11.34DD836 pKa = 3.58LTFSAFVKK844 pKa = 10.61QEE846 pKa = 3.72LLAGYY851 pKa = 10.04KK852 pKa = 10.19KK853 pKa = 10.76FGWADD858 pKa = 3.61AKK860 pKa = 10.75PLAEE864 pKa = 4.85SIARR868 pKa = 11.84MIMAPKK874 pKa = 10.23DD875 pKa = 3.36KK876 pKa = 10.78AHH878 pKa = 6.46IVAGPVIKK886 pKa = 10.4PKK888 pKa = 10.76LEE890 pKa = 3.56RR891 pKa = 11.84LKK893 pKa = 10.92RR894 pKa = 11.84HH895 pKa = 6.15WGPDD899 pKa = 2.42NWLFYY904 pKa = 11.12GATTPEE910 pKa = 4.63KK911 pKa = 10.04LQSWLDD917 pKa = 3.55SCITGCEE924 pKa = 4.04DD925 pKa = 3.94GEE927 pKa = 5.23VFAFWCDD934 pKa = 3.29YY935 pKa = 11.96SMFDD939 pKa = 3.92CTHH942 pKa = 6.13SAEE945 pKa = 4.18SMEE948 pKa = 4.29LVEE951 pKa = 4.94SFYY954 pKa = 11.32SEE956 pKa = 3.91MRR958 pKa = 11.84TDD960 pKa = 3.48PEE962 pKa = 3.77FARR965 pKa = 11.84LINAWRR971 pKa = 11.84APRR974 pKa = 11.84GKK976 pKa = 9.44MGEE979 pKa = 3.92LSYY982 pKa = 10.82RR983 pKa = 11.84AAIMLASGRR992 pKa = 11.84DD993 pKa = 3.57DD994 pKa = 3.51TSLMNAILNGLVMGLSVAAAVAGVEE1019 pKa = 4.16LEE1021 pKa = 4.26SLQKK1025 pKa = 9.97EE1026 pKa = 4.3HH1027 pKa = 7.02LRR1029 pKa = 11.84FAEE1032 pKa = 4.3AYY1034 pKa = 9.26IRR1036 pKa = 11.84ISICGDD1042 pKa = 3.11DD1043 pKa = 3.78TLGFLPKK1050 pKa = 10.28HH1051 pKa = 6.06LWPDD1055 pKa = 3.38RR1056 pKa = 11.84ARR1058 pKa = 11.84IMRR1061 pKa = 11.84DD1062 pKa = 2.45IQTNIARR1069 pKa = 11.84FGLVPKK1075 pKa = 10.39LDD1077 pKa = 3.77CTNFLGSAVYY1087 pKa = 10.53LGMRR1091 pKa = 11.84PYY1093 pKa = 10.87DD1094 pKa = 3.45VPTPTGRR1101 pKa = 11.84KK1102 pKa = 7.14WLWGRR1107 pKa = 11.84TVGRR1111 pKa = 11.84AAYY1114 pKa = 9.46KK1115 pKa = 9.79FGWMLDD1121 pKa = 3.61PSKK1124 pKa = 11.33GDD1126 pKa = 3.31AAAWATGVADD1136 pKa = 3.64SVVRR1140 pKa = 11.84TQPYY1144 pKa = 9.86VPILSDD1150 pKa = 4.56LARR1153 pKa = 11.84QTLKK1157 pKa = 10.64LRR1159 pKa = 11.84EE1160 pKa = 4.18GARR1163 pKa = 11.84RR1164 pKa = 11.84TPVAFEE1170 pKa = 4.19AGKK1173 pKa = 9.1PWTHH1177 pKa = 3.94WTVRR1181 pKa = 11.84EE1182 pKa = 4.04EE1183 pKa = 4.35IKK1185 pKa = 11.06NLTYY1189 pKa = 10.49DD1190 pKa = 3.34RR1191 pKa = 11.84KK1192 pKa = 8.95TIEE1195 pKa = 4.59ALVKK1199 pKa = 10.49SYY1201 pKa = 8.65STPTLYY1207 pKa = 11.04GGDD1210 pKa = 3.8EE1211 pKa = 4.78PICVPSVEE1219 pKa = 4.36DD1220 pKa = 4.13FYY1222 pKa = 11.44RR1223 pKa = 11.84TIATIQAIPCLPYY1236 pKa = 10.94VIDD1239 pKa = 4.7DD1240 pKa = 3.76LALRR1244 pKa = 11.84FMCVTDD1250 pKa = 3.72DD1251 pKa = 3.77CC1252 pKa = 6.43
MM1 pKa = 7.92SITTHH6 pKa = 6.06KK7 pKa = 9.61KK8 pKa = 9.53KK9 pKa = 10.56CGSHH13 pKa = 6.81PDD15 pKa = 3.41VKK17 pKa = 10.64RR18 pKa = 11.84GAEE21 pKa = 3.5QVLFQKK27 pKa = 10.71YY28 pKa = 7.75HH29 pKa = 4.84GKK31 pKa = 9.9YY32 pKa = 8.36GRR34 pKa = 11.84SRR36 pKa = 11.84RR37 pKa = 11.84SYY39 pKa = 10.02LKK41 pKa = 10.54RR42 pKa = 11.84GFGPDD47 pKa = 3.42GGPDD51 pKa = 2.76RR52 pKa = 11.84AVALVHH58 pKa = 6.71RR59 pKa = 11.84LYY61 pKa = 11.09AGLRR65 pKa = 11.84GAGRR69 pKa = 11.84YY70 pKa = 7.52PVPQNPSDD78 pKa = 3.71ILNHH82 pKa = 6.4LLATNVIDD90 pKa = 3.84GRR92 pKa = 11.84YY93 pKa = 9.02RR94 pKa = 11.84DD95 pKa = 3.92EE96 pKa = 4.36EE97 pKa = 4.22KK98 pKa = 11.05GGLGLPCSLTAFATAFPNQRR118 pKa = 11.84HH119 pKa = 5.35ICLFYY124 pKa = 11.14EE125 pKa = 4.46KK126 pKa = 10.51LCEE129 pKa = 4.22QGLHH133 pKa = 6.04EE134 pKa = 4.89EE135 pKa = 4.3NSRR138 pKa = 11.84GPSGGMTLTGMVSALQEE155 pKa = 3.89WEE157 pKa = 3.94AGIIADD163 pKa = 4.52PKK165 pKa = 9.39PKK167 pKa = 9.98VVRR170 pKa = 11.84PRR172 pKa = 11.84GMDD175 pKa = 3.32LHH177 pKa = 6.48QVNCRR182 pKa = 11.84GGTFLSYY189 pKa = 11.03DD190 pKa = 3.62VVHH193 pKa = 6.19RR194 pKa = 11.84TLLRR198 pKa = 11.84DD199 pKa = 3.49GARR202 pKa = 11.84EE203 pKa = 3.79QIIVYY208 pKa = 10.39INDD211 pKa = 3.16EE212 pKa = 4.3SCNFGHH218 pKa = 6.62FCAVSSLNEE227 pKa = 3.76DD228 pKa = 2.95AMKK231 pKa = 10.81RR232 pKa = 11.84EE233 pKa = 3.66IAKK236 pKa = 9.79GKK238 pKa = 10.06RR239 pKa = 11.84RR240 pKa = 11.84AEE242 pKa = 4.11YY243 pKa = 10.89ADD245 pKa = 3.64MVAADD250 pKa = 3.95SDD252 pKa = 4.09QVPKK256 pKa = 10.6EE257 pKa = 4.11VAPKK261 pKa = 10.23AISGDD266 pKa = 3.92HH267 pKa = 6.14IVPATTSEE275 pKa = 4.27PGPSGINQEE284 pKa = 4.48AEE286 pKa = 4.08THH288 pKa = 6.11HH289 pKa = 6.23SQEE292 pKa = 3.72LDD294 pKa = 3.26ALLEE298 pKa = 3.92EE299 pKa = 5.93AIAEE303 pKa = 4.25MQGMDD308 pKa = 4.51PPQPPPPEE316 pKa = 4.82PNDD319 pKa = 3.4NGPGGNIGGEE329 pKa = 4.03GDD331 pKa = 4.52GPDD334 pKa = 3.69EE335 pKa = 4.14PPIIKK340 pKa = 10.34EE341 pKa = 4.14EE342 pKa = 4.24VPPPPPPPPPEE353 pKa = 4.25PKK355 pKa = 9.75PIPVSRR361 pKa = 11.84RR362 pKa = 11.84TLRR365 pKa = 11.84YY366 pKa = 9.38LRR368 pKa = 11.84EE369 pKa = 3.67KK370 pKa = 11.23LEE372 pKa = 4.46FIKK375 pKa = 9.33THH377 pKa = 5.83SATPILDD384 pKa = 3.56HH385 pKa = 6.82RR386 pKa = 11.84PVRR389 pKa = 11.84AYY391 pKa = 10.32ACAGLVSEE399 pKa = 5.09GCVEE403 pKa = 4.49HH404 pKa = 7.39EE405 pKa = 4.51DD406 pKa = 3.78ASHH409 pKa = 6.4EE410 pKa = 4.34GGVGLTEE417 pKa = 3.97LHH419 pKa = 6.75PEE421 pKa = 3.65VLARR425 pKa = 11.84FNFTRR430 pKa = 11.84RR431 pKa = 11.84SYY433 pKa = 10.35YY434 pKa = 9.78YY435 pKa = 8.23VTAGVEE441 pKa = 3.92PDD443 pKa = 3.47ALDD446 pKa = 4.56GRR448 pKa = 11.84FLTNGQYY455 pKa = 10.56NPAWVASIQFEE466 pKa = 4.31DD467 pKa = 3.75TAFTLEE473 pKa = 4.01FVGNKK478 pKa = 9.61GPCDD482 pKa = 3.5YY483 pKa = 11.19YY484 pKa = 11.15RR485 pKa = 11.84LYY487 pKa = 10.83KK488 pKa = 10.57AATLGAKK495 pKa = 9.79KK496 pKa = 10.64GFFSCPDD503 pKa = 4.68FINEE507 pKa = 4.18ALDD510 pKa = 5.4DD511 pKa = 4.09VSAWAAEE518 pKa = 4.12IGLITQPQATIKK530 pKa = 10.59HH531 pKa = 4.66VTNLPGRR538 pKa = 11.84GLRR541 pKa = 11.84PRR543 pKa = 11.84SLKK546 pKa = 10.46GVAGASRR553 pKa = 11.84AQWAMHH559 pKa = 6.24ARR561 pKa = 11.84HH562 pKa = 6.25APEE565 pKa = 4.66DD566 pKa = 3.51LRR568 pKa = 11.84GPYY571 pKa = 9.82HH572 pKa = 7.15SLFIEE577 pKa = 4.37NADD580 pKa = 4.96DD581 pKa = 4.69SNSDD585 pKa = 3.55PNEE588 pKa = 3.57VLKK591 pKa = 10.88ALKK594 pKa = 10.1YY595 pKa = 9.47EE596 pKa = 3.87KK597 pKa = 11.29AMMEE601 pKa = 4.15KK602 pKa = 9.68EE603 pKa = 3.87HH604 pKa = 6.64GKK606 pKa = 10.2RR607 pKa = 11.84GYY609 pKa = 9.57CAPKK613 pKa = 9.08MSTPKK618 pKa = 9.27TCASCGVSPPKK629 pKa = 10.57GKK631 pKa = 10.71YY632 pKa = 8.04KK633 pKa = 9.94WKK635 pKa = 10.1HH636 pKa = 4.92RR637 pKa = 11.84QCKK640 pKa = 10.1DD641 pKa = 3.2CQQKK645 pKa = 10.8LKK647 pKa = 9.71TCGAVSTMGQQIQQNLTVAEE667 pKa = 4.07GHH669 pKa = 6.83PGCVHH674 pKa = 6.74LNSSTLPPKK683 pKa = 9.87KK684 pKa = 10.15EE685 pKa = 3.47KK686 pKa = 8.47WAKK689 pKa = 8.66VHH691 pKa = 6.11IPKK694 pKa = 10.47GAIKK698 pKa = 9.75VHH700 pKa = 6.44KK701 pKa = 10.43SDD703 pKa = 4.53VPWLRR708 pKa = 11.84HH709 pKa = 5.65ADD711 pKa = 3.12WGGSKK716 pKa = 8.22MHH718 pKa = 6.24SVEE721 pKa = 5.46KK722 pKa = 10.55EE723 pKa = 3.92DD724 pKa = 4.56LSKK727 pKa = 10.54IDD729 pKa = 3.61TTLEE733 pKa = 3.81RR734 pKa = 11.84PKK736 pKa = 10.46RR737 pKa = 11.84EE738 pKa = 3.98CVLAGIGVSGCYY750 pKa = 10.11PMVTRR755 pKa = 11.84KK756 pKa = 10.05GFYY759 pKa = 10.85ARR761 pKa = 11.84MQALIGRR768 pKa = 11.84AYY770 pKa = 9.78LQKK773 pKa = 10.52PEE775 pKa = 4.29SSPAAWEE782 pKa = 4.48AMNQMKK788 pKa = 10.47DD789 pKa = 3.36CLLPNDD795 pKa = 4.64ALDD798 pKa = 4.09GEE800 pKa = 4.68KK801 pKa = 10.7FSIDD805 pKa = 3.12FWLTTMPARR814 pKa = 11.84RR815 pKa = 11.84RR816 pKa = 11.84RR817 pKa = 11.84ALEE820 pKa = 4.04RR821 pKa = 11.84AHH823 pKa = 7.26KK824 pKa = 9.64EE825 pKa = 3.97YY826 pKa = 10.76KK827 pKa = 10.27EE828 pKa = 4.16SGGLRR833 pKa = 11.84DD834 pKa = 3.79KK835 pKa = 11.34DD836 pKa = 3.58LTFSAFVKK844 pKa = 10.61QEE846 pKa = 3.72LLAGYY851 pKa = 10.04KK852 pKa = 10.19KK853 pKa = 10.76FGWADD858 pKa = 3.61AKK860 pKa = 10.75PLAEE864 pKa = 4.85SIARR868 pKa = 11.84MIMAPKK874 pKa = 10.23DD875 pKa = 3.36KK876 pKa = 10.78AHH878 pKa = 6.46IVAGPVIKK886 pKa = 10.4PKK888 pKa = 10.76LEE890 pKa = 3.56RR891 pKa = 11.84LKK893 pKa = 10.92RR894 pKa = 11.84HH895 pKa = 6.15WGPDD899 pKa = 2.42NWLFYY904 pKa = 11.12GATTPEE910 pKa = 4.63KK911 pKa = 10.04LQSWLDD917 pKa = 3.55SCITGCEE924 pKa = 4.04DD925 pKa = 3.94GEE927 pKa = 5.23VFAFWCDD934 pKa = 3.29YY935 pKa = 11.96SMFDD939 pKa = 3.92CTHH942 pKa = 6.13SAEE945 pKa = 4.18SMEE948 pKa = 4.29LVEE951 pKa = 4.94SFYY954 pKa = 11.32SEE956 pKa = 3.91MRR958 pKa = 11.84TDD960 pKa = 3.48PEE962 pKa = 3.77FARR965 pKa = 11.84LINAWRR971 pKa = 11.84APRR974 pKa = 11.84GKK976 pKa = 9.44MGEE979 pKa = 3.92LSYY982 pKa = 10.82RR983 pKa = 11.84AAIMLASGRR992 pKa = 11.84DD993 pKa = 3.57DD994 pKa = 3.51TSLMNAILNGLVMGLSVAAAVAGVEE1019 pKa = 4.16LEE1021 pKa = 4.26SLQKK1025 pKa = 9.97EE1026 pKa = 4.3HH1027 pKa = 7.02LRR1029 pKa = 11.84FAEE1032 pKa = 4.3AYY1034 pKa = 9.26IRR1036 pKa = 11.84ISICGDD1042 pKa = 3.11DD1043 pKa = 3.78TLGFLPKK1050 pKa = 10.28HH1051 pKa = 6.06LWPDD1055 pKa = 3.38RR1056 pKa = 11.84ARR1058 pKa = 11.84IMRR1061 pKa = 11.84DD1062 pKa = 2.45IQTNIARR1069 pKa = 11.84FGLVPKK1075 pKa = 10.39LDD1077 pKa = 3.77CTNFLGSAVYY1087 pKa = 10.53LGMRR1091 pKa = 11.84PYY1093 pKa = 10.87DD1094 pKa = 3.45VPTPTGRR1101 pKa = 11.84KK1102 pKa = 7.14WLWGRR1107 pKa = 11.84TVGRR1111 pKa = 11.84AAYY1114 pKa = 9.46KK1115 pKa = 9.79FGWMLDD1121 pKa = 3.61PSKK1124 pKa = 11.33GDD1126 pKa = 3.31AAAWATGVADD1136 pKa = 3.64SVVRR1140 pKa = 11.84TQPYY1144 pKa = 9.86VPILSDD1150 pKa = 4.56LARR1153 pKa = 11.84QTLKK1157 pKa = 10.64LRR1159 pKa = 11.84EE1160 pKa = 4.18GARR1163 pKa = 11.84RR1164 pKa = 11.84TPVAFEE1170 pKa = 4.19AGKK1173 pKa = 9.1PWTHH1177 pKa = 3.94WTVRR1181 pKa = 11.84EE1182 pKa = 4.04EE1183 pKa = 4.35IKK1185 pKa = 11.06NLTYY1189 pKa = 10.49DD1190 pKa = 3.34RR1191 pKa = 11.84KK1192 pKa = 8.95TIEE1195 pKa = 4.59ALVKK1199 pKa = 10.49SYY1201 pKa = 8.65STPTLYY1207 pKa = 11.04GGDD1210 pKa = 3.8EE1211 pKa = 4.78PICVPSVEE1219 pKa = 4.36DD1220 pKa = 4.13FYY1222 pKa = 11.44RR1223 pKa = 11.84TIATIQAIPCLPYY1236 pKa = 10.94VIDD1239 pKa = 4.7DD1240 pKa = 3.76LALRR1244 pKa = 11.84FMCVTDD1250 pKa = 3.72DD1251 pKa = 3.77CC1252 pKa = 6.43
Molecular weight: 139.03 kDa
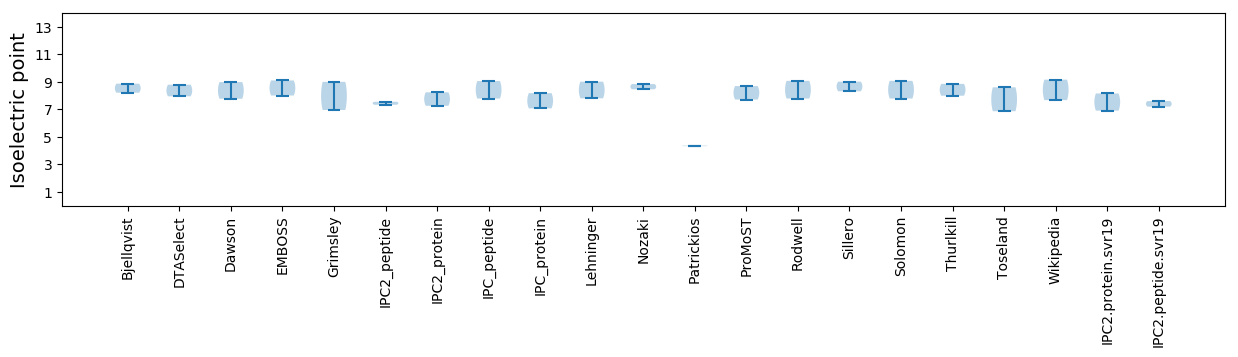
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLD2|A0A1L3KLD2_9VIRU Uncharacterized protein OS=Wenzhou yanvirus-like virus 2 OX=1923684 PE=4 SV=1
MM1 pKa = 7.35SHH3 pKa = 6.77TGLKK7 pKa = 10.42SLDD10 pKa = 4.15EE11 pKa = 4.22IAQTICLPNEE21 pKa = 4.01RR22 pKa = 11.84APVRR26 pKa = 11.84LPTYY30 pKa = 10.14PSIDD34 pKa = 3.02KK35 pKa = 9.25TATFRR40 pKa = 11.84YY41 pKa = 9.7RR42 pKa = 11.84YY43 pKa = 10.05QNTQSLRR50 pKa = 11.84DD51 pKa = 3.49EE52 pKa = 5.29ADD54 pKa = 3.53PSDD57 pKa = 3.84QNVLGRR63 pKa = 11.84KK64 pKa = 9.0RR65 pKa = 11.84FLVTRR70 pKa = 11.84DD71 pKa = 3.21PAAPLLLDD79 pKa = 4.09TVHH82 pKa = 6.96TLQKK86 pKa = 10.3AISGNPSIYY95 pKa = 9.67TGSVLARR102 pKa = 11.84TEE104 pKa = 3.86PVNINTADD112 pKa = 3.84EE113 pKa = 4.39LTTATVTLSKK123 pKa = 9.89MYY125 pKa = 9.57PWEE128 pKa = 4.37FYY130 pKa = 10.84NRR132 pKa = 11.84LPSGEE137 pKa = 4.32YY138 pKa = 9.95DD139 pKa = 3.3SKK141 pKa = 11.37QWFVVPKK148 pKa = 9.69TLDD151 pKa = 3.26SYY153 pKa = 11.72GRR155 pKa = 11.84ASGFLGHH162 pKa = 7.28LGVALFVTDD171 pKa = 5.07IAPQTQPIVTYY182 pKa = 10.52KK183 pKa = 10.68PGDD186 pKa = 3.76PVSEE190 pKa = 4.2IPVGALVLDD199 pKa = 3.77YY200 pKa = 10.29TLTIEE205 pKa = 4.58IVDD208 pKa = 3.89STGVAGSMICDD219 pKa = 4.05LDD221 pKa = 3.74YY222 pKa = 11.27TKK224 pKa = 10.44TRR226 pKa = 11.84EE227 pKa = 4.19APGTYY232 pKa = 9.13TGFFIIDD239 pKa = 3.4PNAALVRR246 pKa = 11.84IQSLSYY252 pKa = 9.85TGPIKK257 pKa = 10.23MKK259 pKa = 10.31QSNGTLTVIPEE270 pKa = 4.11RR271 pKa = 11.84NVYY274 pKa = 8.23PTILLTEE281 pKa = 4.75KK282 pKa = 10.6GSHH285 pKa = 6.6PPGATKK291 pKa = 9.66TFRR294 pKa = 11.84CLSYY298 pKa = 10.17PATSVNPEE306 pKa = 4.2YY307 pKa = 10.47YY308 pKa = 10.6NSVAPFRR315 pKa = 11.84STRR318 pKa = 11.84LNASALLLTNVTKK331 pKa = 10.79VLNKK335 pKa = 9.91EE336 pKa = 4.13GTVEE340 pKa = 3.9SSRR343 pKa = 11.84VIFNPDD349 pKa = 2.72SGRR352 pKa = 11.84TFMNADD358 pKa = 3.49VPTVSTSNPEE368 pKa = 3.47TRR370 pKa = 11.84YY371 pKa = 10.25FGALEE376 pKa = 3.95KK377 pKa = 10.7GAYY380 pKa = 8.23TFTAPDD386 pKa = 3.4QEE388 pKa = 4.64SLVFTTPYY396 pKa = 8.39EE397 pKa = 4.25TVRR400 pKa = 11.84VNDD403 pKa = 3.81ALIGHH408 pKa = 7.35GGGSVPIFVPEE419 pKa = 3.98RR420 pKa = 11.84LAEE423 pKa = 4.01RR424 pKa = 11.84PVMNLNPRR432 pKa = 11.84YY433 pKa = 8.66MNAIILTDD441 pKa = 4.31LDD443 pKa = 4.03MTDD446 pKa = 3.83DD447 pKa = 3.75TQLALTLDD455 pKa = 3.78THH457 pKa = 5.87WEE459 pKa = 3.82FRR461 pKa = 11.84TISTLYY467 pKa = 9.4QLDD470 pKa = 4.11YY471 pKa = 11.39SRR473 pKa = 11.84MPIEE477 pKa = 4.51TYY479 pKa = 9.85HH480 pKa = 7.02AAMLAVMRR488 pKa = 11.84AGLFYY493 pKa = 10.94EE494 pKa = 4.59NNTHH498 pKa = 6.48QRR500 pKa = 11.84ILQAVGKK507 pKa = 8.47GLKK510 pKa = 9.32FAAPLIPGGNMLQLAHH526 pKa = 6.55SAAKK530 pKa = 10.11AVADD534 pKa = 3.6SRR536 pKa = 11.84GKK538 pKa = 10.69KK539 pKa = 8.41KK540 pKa = 9.19TPTANSSKK548 pKa = 9.36RR549 pKa = 11.84TKK551 pKa = 10.33DD552 pKa = 3.14SKK554 pKa = 8.14QTKK557 pKa = 10.21GSMKK561 pKa = 10.46QKK563 pKa = 10.47GLKK566 pKa = 9.49
MM1 pKa = 7.35SHH3 pKa = 6.77TGLKK7 pKa = 10.42SLDD10 pKa = 4.15EE11 pKa = 4.22IAQTICLPNEE21 pKa = 4.01RR22 pKa = 11.84APVRR26 pKa = 11.84LPTYY30 pKa = 10.14PSIDD34 pKa = 3.02KK35 pKa = 9.25TATFRR40 pKa = 11.84YY41 pKa = 9.7RR42 pKa = 11.84YY43 pKa = 10.05QNTQSLRR50 pKa = 11.84DD51 pKa = 3.49EE52 pKa = 5.29ADD54 pKa = 3.53PSDD57 pKa = 3.84QNVLGRR63 pKa = 11.84KK64 pKa = 9.0RR65 pKa = 11.84FLVTRR70 pKa = 11.84DD71 pKa = 3.21PAAPLLLDD79 pKa = 4.09TVHH82 pKa = 6.96TLQKK86 pKa = 10.3AISGNPSIYY95 pKa = 9.67TGSVLARR102 pKa = 11.84TEE104 pKa = 3.86PVNINTADD112 pKa = 3.84EE113 pKa = 4.39LTTATVTLSKK123 pKa = 9.89MYY125 pKa = 9.57PWEE128 pKa = 4.37FYY130 pKa = 10.84NRR132 pKa = 11.84LPSGEE137 pKa = 4.32YY138 pKa = 9.95DD139 pKa = 3.3SKK141 pKa = 11.37QWFVVPKK148 pKa = 9.69TLDD151 pKa = 3.26SYY153 pKa = 11.72GRR155 pKa = 11.84ASGFLGHH162 pKa = 7.28LGVALFVTDD171 pKa = 5.07IAPQTQPIVTYY182 pKa = 10.52KK183 pKa = 10.68PGDD186 pKa = 3.76PVSEE190 pKa = 4.2IPVGALVLDD199 pKa = 3.77YY200 pKa = 10.29TLTIEE205 pKa = 4.58IVDD208 pKa = 3.89STGVAGSMICDD219 pKa = 4.05LDD221 pKa = 3.74YY222 pKa = 11.27TKK224 pKa = 10.44TRR226 pKa = 11.84EE227 pKa = 4.19APGTYY232 pKa = 9.13TGFFIIDD239 pKa = 3.4PNAALVRR246 pKa = 11.84IQSLSYY252 pKa = 9.85TGPIKK257 pKa = 10.23MKK259 pKa = 10.31QSNGTLTVIPEE270 pKa = 4.11RR271 pKa = 11.84NVYY274 pKa = 8.23PTILLTEE281 pKa = 4.75KK282 pKa = 10.6GSHH285 pKa = 6.6PPGATKK291 pKa = 9.66TFRR294 pKa = 11.84CLSYY298 pKa = 10.17PATSVNPEE306 pKa = 4.2YY307 pKa = 10.47YY308 pKa = 10.6NSVAPFRR315 pKa = 11.84STRR318 pKa = 11.84LNASALLLTNVTKK331 pKa = 10.79VLNKK335 pKa = 9.91EE336 pKa = 4.13GTVEE340 pKa = 3.9SSRR343 pKa = 11.84VIFNPDD349 pKa = 2.72SGRR352 pKa = 11.84TFMNADD358 pKa = 3.49VPTVSTSNPEE368 pKa = 3.47TRR370 pKa = 11.84YY371 pKa = 10.25FGALEE376 pKa = 3.95KK377 pKa = 10.7GAYY380 pKa = 8.23TFTAPDD386 pKa = 3.4QEE388 pKa = 4.64SLVFTTPYY396 pKa = 8.39EE397 pKa = 4.25TVRR400 pKa = 11.84VNDD403 pKa = 3.81ALIGHH408 pKa = 7.35GGGSVPIFVPEE419 pKa = 3.98RR420 pKa = 11.84LAEE423 pKa = 4.01RR424 pKa = 11.84PVMNLNPRR432 pKa = 11.84YY433 pKa = 8.66MNAIILTDD441 pKa = 4.31LDD443 pKa = 4.03MTDD446 pKa = 3.83DD447 pKa = 3.75TQLALTLDD455 pKa = 3.78THH457 pKa = 5.87WEE459 pKa = 3.82FRR461 pKa = 11.84TISTLYY467 pKa = 9.4QLDD470 pKa = 4.11YY471 pKa = 11.39SRR473 pKa = 11.84MPIEE477 pKa = 4.51TYY479 pKa = 9.85HH480 pKa = 7.02AAMLAVMRR488 pKa = 11.84AGLFYY493 pKa = 10.94EE494 pKa = 4.59NNTHH498 pKa = 6.48QRR500 pKa = 11.84ILQAVGKK507 pKa = 8.47GLKK510 pKa = 9.32FAAPLIPGGNMLQLAHH526 pKa = 6.55SAAKK530 pKa = 10.11AVADD534 pKa = 3.6SRR536 pKa = 11.84GKK538 pKa = 10.69KK539 pKa = 8.41KK540 pKa = 9.19TPTANSSKK548 pKa = 9.36RR549 pKa = 11.84TKK551 pKa = 10.33DD552 pKa = 3.14SKK554 pKa = 8.14QTKK557 pKa = 10.21GSMKK561 pKa = 10.46QKK563 pKa = 10.47GLKK566 pKa = 9.49
Molecular weight: 62.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1818 |
566 |
1252 |
909.0 |
100.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.801 ± 0.552 | 1.87 ± 0.72 |
5.556 ± 0.232 | 5.831 ± 0.855 |
3.19 ± 0.09 | 7.591 ± 0.756 |
2.53 ± 0.505 | 4.51 ± 0.14 |
5.996 ± 0.469 | 8.526 ± 0.545 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.475 ± 0.096 | 3.245 ± 0.725 |
7.261 ± 0.104 | 2.915 ± 0.142 |
6.106 ± 0.433 | 5.941 ± 0.605 |
6.766 ± 2.156 | 5.776 ± 0.409 |
1.43 ± 0.484 | 3.685 ± 0.393 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
