
Ashy storm petrel gyrovirus
Taxonomy: Viruses; Anelloviridae; Gyrovirus; Gyrovirus hydho1
Average proteome isoelectric point is 8.41
Get precalculated fractions of proteins
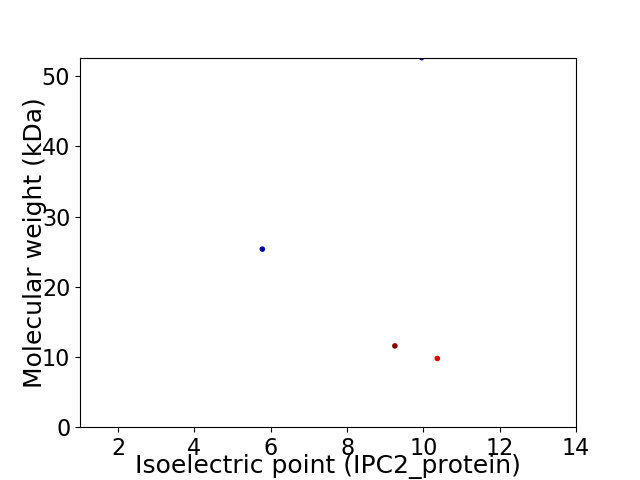
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z4N430|A0A2Z4N430_9VIRU CA1 OS=Ashy storm petrel gyrovirus OX=2249930 PE=3 SV=1
MM1 pKa = 7.31SFSVLCRR8 pKa = 11.84DD9 pKa = 3.86TLRR12 pKa = 11.84GTLQYY17 pKa = 10.51NPKK20 pKa = 9.1YY21 pKa = 10.53QRR23 pKa = 11.84DD24 pKa = 3.41LANWFRR30 pKa = 11.84DD31 pKa = 3.42CSRR34 pKa = 11.84THH36 pKa = 7.06RR37 pKa = 11.84AICRR41 pKa = 11.84CPSYY45 pKa = 10.62RR46 pKa = 11.84NHH48 pKa = 6.55FKK50 pKa = 10.81APKK53 pKa = 8.97PQTQEE58 pKa = 4.0ASTQTEE64 pKa = 4.23RR65 pKa = 11.84PPKK68 pKa = 10.36SVTILPARR76 pKa = 11.84RR77 pKa = 11.84GILSSPKK84 pKa = 9.86DD85 pKa = 3.52RR86 pKa = 11.84SPGKK90 pKa = 9.91RR91 pKa = 11.84NKK93 pKa = 9.52ILHH96 pKa = 6.75FDD98 pKa = 3.68PFLNEE103 pKa = 3.85ALYY106 pKa = 10.35HH107 pKa = 5.3SKK109 pKa = 10.38RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84KK113 pKa = 9.39QLLQEE118 pKa = 5.17RR119 pKa = 11.84PDD121 pKa = 3.16NWHH124 pKa = 6.93PLWEE128 pKa = 4.14ITPPLKK134 pKa = 10.78ACLDD138 pKa = 3.79FDD140 pKa = 4.19DD141 pKa = 5.02TEE143 pKa = 4.59EE144 pKa = 4.55DD145 pKa = 3.82TDD147 pKa = 4.1TSDD150 pKa = 4.34DD151 pKa = 3.95DD152 pKa = 4.58GGAGEE157 pKa = 5.13TDD159 pKa = 4.27DD160 pKa = 5.6EE161 pKa = 6.15DD162 pKa = 3.87DD163 pKa = 4.14TVPHH167 pKa = 6.46VDD169 pKa = 4.25FDD171 pKa = 3.57IGINGIVDD179 pKa = 5.14GIPPEE184 pKa = 4.38PSDD187 pKa = 3.63ASSSNRR193 pKa = 11.84TLEE196 pKa = 3.92QNPYY200 pKa = 10.25GLPNHH205 pKa = 5.74TTHH208 pKa = 7.15SEE210 pKa = 4.05LLSKK214 pKa = 10.82VLFSSHH220 pKa = 5.35KK221 pKa = 9.24QQ222 pKa = 3.17
MM1 pKa = 7.31SFSVLCRR8 pKa = 11.84DD9 pKa = 3.86TLRR12 pKa = 11.84GTLQYY17 pKa = 10.51NPKK20 pKa = 9.1YY21 pKa = 10.53QRR23 pKa = 11.84DD24 pKa = 3.41LANWFRR30 pKa = 11.84DD31 pKa = 3.42CSRR34 pKa = 11.84THH36 pKa = 7.06RR37 pKa = 11.84AICRR41 pKa = 11.84CPSYY45 pKa = 10.62RR46 pKa = 11.84NHH48 pKa = 6.55FKK50 pKa = 10.81APKK53 pKa = 8.97PQTQEE58 pKa = 4.0ASTQTEE64 pKa = 4.23RR65 pKa = 11.84PPKK68 pKa = 10.36SVTILPARR76 pKa = 11.84RR77 pKa = 11.84GILSSPKK84 pKa = 9.86DD85 pKa = 3.52RR86 pKa = 11.84SPGKK90 pKa = 9.91RR91 pKa = 11.84NKK93 pKa = 9.52ILHH96 pKa = 6.75FDD98 pKa = 3.68PFLNEE103 pKa = 3.85ALYY106 pKa = 10.35HH107 pKa = 5.3SKK109 pKa = 10.38RR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84KK113 pKa = 9.39QLLQEE118 pKa = 5.17RR119 pKa = 11.84PDD121 pKa = 3.16NWHH124 pKa = 6.93PLWEE128 pKa = 4.14ITPPLKK134 pKa = 10.78ACLDD138 pKa = 3.79FDD140 pKa = 4.19DD141 pKa = 5.02TEE143 pKa = 4.59EE144 pKa = 4.55DD145 pKa = 3.82TDD147 pKa = 4.1TSDD150 pKa = 4.34DD151 pKa = 3.95DD152 pKa = 4.58GGAGEE157 pKa = 5.13TDD159 pKa = 4.27DD160 pKa = 5.6EE161 pKa = 6.15DD162 pKa = 3.87DD163 pKa = 4.14TVPHH167 pKa = 6.46VDD169 pKa = 4.25FDD171 pKa = 3.57IGINGIVDD179 pKa = 5.14GIPPEE184 pKa = 4.38PSDD187 pKa = 3.63ASSSNRR193 pKa = 11.84TLEE196 pKa = 3.92QNPYY200 pKa = 10.25GLPNHH205 pKa = 5.74TTHH208 pKa = 7.15SEE210 pKa = 4.05LLSKK214 pKa = 10.82VLFSSHH220 pKa = 5.35KK221 pKa = 9.24QQ222 pKa = 3.17
Molecular weight: 25.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z4N434|A0A2Z4N434_9VIRU ORF3 OS=Ashy storm petrel gyrovirus OX=2249930 PE=4 SV=1
MM1 pKa = 7.41SRR3 pKa = 11.84FRR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 9.48RR8 pKa = 11.84GRR10 pKa = 11.84YY11 pKa = 7.71GYY13 pKa = 10.45LGRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84WRR20 pKa = 11.84WRR22 pKa = 11.84NRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.43RR30 pKa = 11.84PTRR33 pKa = 11.84RR34 pKa = 11.84LRR36 pKa = 11.84YY37 pKa = 8.22RR38 pKa = 11.84YY39 pKa = 9.59KK40 pKa = 10.2RR41 pKa = 11.84YY42 pKa = 9.79RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84NSARR49 pKa = 11.84AIRR52 pKa = 11.84RR53 pKa = 11.84LFFKK57 pKa = 10.03PNPGTEE63 pKa = 4.14PVRR66 pKa = 11.84ITKK69 pKa = 9.79PYY71 pKa = 9.66NSLGVTFQGIILIPQTVAAWSLNVPQDD98 pKa = 3.16QTDD101 pKa = 3.37NYY103 pKa = 9.97ISSRR107 pKa = 11.84VAKK110 pKa = 10.09IDD112 pKa = 3.37VSFANLLRR120 pKa = 11.84AVLPLDD126 pKa = 4.27YY127 pKa = 9.85ISKK130 pKa = 10.18IGGPIPGRR138 pKa = 11.84LQSQVTGQPSDD149 pKa = 3.6TPPPKK154 pKa = 10.24TGWPYY159 pKa = 11.17AKK161 pKa = 10.38PMTQDD166 pKa = 2.92GQTPTSNPSEE176 pKa = 3.83WWRR179 pKa = 11.84WALLLMHH186 pKa = 6.9PTDD189 pKa = 3.3NVRR192 pKa = 11.84FLTTPQIMNTEE203 pKa = 3.84EE204 pKa = 3.94MGAMFAQYY212 pKa = 10.61QLLRR216 pKa = 11.84HH217 pKa = 5.94EE218 pKa = 4.45FTRR221 pKa = 11.84FKK223 pKa = 11.11VEE225 pKa = 4.59CIDD228 pKa = 4.95DD229 pKa = 5.02GGLKK233 pKa = 9.58WSPVASLAIQNQYY246 pKa = 9.83WEE248 pKa = 4.62IKK250 pKa = 10.25SNPTSPYY257 pKa = 7.29TAHH260 pKa = 7.01TIVQPGTQTEE270 pKa = 4.4IVNQATGPRR279 pKa = 11.84TPTDD283 pKa = 3.13QDD285 pKa = 3.76CPTRR289 pKa = 11.84QYY291 pKa = 11.33PRR293 pKa = 11.84YY294 pKa = 8.71LHH296 pKa = 6.72NGQTPDD302 pKa = 3.18KK303 pKa = 10.16LANTFSTILCKK314 pKa = 9.41EE315 pKa = 3.87TGYY318 pKa = 10.4IFPSFATLSAMGAPWAFPPQQKK340 pKa = 9.67PVGRR344 pKa = 11.84GSFNKK349 pKa = 10.14HH350 pKa = 4.65KK351 pKa = 10.43MKK353 pKa = 10.6GRR355 pKa = 11.84NDD357 pKa = 3.89PLKK360 pKa = 10.29GPWMTLLPKK369 pKa = 10.32NICNVSEE376 pKa = 4.16NEE378 pKa = 4.18MVGGDD383 pKa = 3.99LNTIIGTLFLAQGSNHH399 pKa = 4.21YY400 pKa = 6.7TTYY403 pKa = 11.57KK404 pKa = 10.12FMTMQDD410 pKa = 3.34TVQPLIMQTRR420 pKa = 11.84AWAAVKK426 pKa = 9.98IQSRR430 pKa = 11.84WTLANTRR437 pKa = 11.84RR438 pKa = 11.84PYY440 pKa = 10.42RR441 pKa = 11.84WDD443 pKa = 3.49VAWSNTTGKK452 pKa = 10.24FPQQ455 pKa = 3.75
MM1 pKa = 7.41SRR3 pKa = 11.84FRR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 9.48RR8 pKa = 11.84GRR10 pKa = 11.84YY11 pKa = 7.71GYY13 pKa = 10.45LGRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84WRR20 pKa = 11.84WRR22 pKa = 11.84NRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.43RR30 pKa = 11.84PTRR33 pKa = 11.84RR34 pKa = 11.84LRR36 pKa = 11.84YY37 pKa = 8.22RR38 pKa = 11.84YY39 pKa = 9.59KK40 pKa = 10.2RR41 pKa = 11.84YY42 pKa = 9.79RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84NSARR49 pKa = 11.84AIRR52 pKa = 11.84RR53 pKa = 11.84LFFKK57 pKa = 10.03PNPGTEE63 pKa = 4.14PVRR66 pKa = 11.84ITKK69 pKa = 9.79PYY71 pKa = 9.66NSLGVTFQGIILIPQTVAAWSLNVPQDD98 pKa = 3.16QTDD101 pKa = 3.37NYY103 pKa = 9.97ISSRR107 pKa = 11.84VAKK110 pKa = 10.09IDD112 pKa = 3.37VSFANLLRR120 pKa = 11.84AVLPLDD126 pKa = 4.27YY127 pKa = 9.85ISKK130 pKa = 10.18IGGPIPGRR138 pKa = 11.84LQSQVTGQPSDD149 pKa = 3.6TPPPKK154 pKa = 10.24TGWPYY159 pKa = 11.17AKK161 pKa = 10.38PMTQDD166 pKa = 2.92GQTPTSNPSEE176 pKa = 3.83WWRR179 pKa = 11.84WALLLMHH186 pKa = 6.9PTDD189 pKa = 3.3NVRR192 pKa = 11.84FLTTPQIMNTEE203 pKa = 3.84EE204 pKa = 3.94MGAMFAQYY212 pKa = 10.61QLLRR216 pKa = 11.84HH217 pKa = 5.94EE218 pKa = 4.45FTRR221 pKa = 11.84FKK223 pKa = 11.11VEE225 pKa = 4.59CIDD228 pKa = 4.95DD229 pKa = 5.02GGLKK233 pKa = 9.58WSPVASLAIQNQYY246 pKa = 9.83WEE248 pKa = 4.62IKK250 pKa = 10.25SNPTSPYY257 pKa = 7.29TAHH260 pKa = 7.01TIVQPGTQTEE270 pKa = 4.4IVNQATGPRR279 pKa = 11.84TPTDD283 pKa = 3.13QDD285 pKa = 3.76CPTRR289 pKa = 11.84QYY291 pKa = 11.33PRR293 pKa = 11.84YY294 pKa = 8.71LHH296 pKa = 6.72NGQTPDD302 pKa = 3.18KK303 pKa = 10.16LANTFSTILCKK314 pKa = 9.41EE315 pKa = 3.87TGYY318 pKa = 10.4IFPSFATLSAMGAPWAFPPQQKK340 pKa = 9.67PVGRR344 pKa = 11.84GSFNKK349 pKa = 10.14HH350 pKa = 4.65KK351 pKa = 10.43MKK353 pKa = 10.6GRR355 pKa = 11.84NDD357 pKa = 3.89PLKK360 pKa = 10.29GPWMTLLPKK369 pKa = 10.32NICNVSEE376 pKa = 4.16NEE378 pKa = 4.18MVGGDD383 pKa = 3.99LNTIIGTLFLAQGSNHH399 pKa = 4.21YY400 pKa = 6.7TTYY403 pKa = 11.57KK404 pKa = 10.12FMTMQDD410 pKa = 3.34TVQPLIMQTRR420 pKa = 11.84AWAAVKK426 pKa = 9.98IQSRR430 pKa = 11.84WTLANTRR437 pKa = 11.84RR438 pKa = 11.84PYY440 pKa = 10.42RR441 pKa = 11.84WDD443 pKa = 3.49VAWSNTTGKK452 pKa = 10.24FPQQ455 pKa = 3.75
Molecular weight: 52.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
869 |
90 |
455 |
217.3 |
24.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.674 ± 1.619 | 1.381 ± 0.387 |
4.948 ± 1.798 | 3.567 ± 0.92 |
3.567 ± 0.311 | 5.639 ± 0.603 |
2.647 ± 0.992 | 4.948 ± 0.348 |
4.488 ± 0.454 | 7.595 ± 0.948 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.956 ± 0.68 | 4.488 ± 0.655 |
8.861 ± 0.453 | 4.948 ± 0.825 |
10.127 ± 1.011 | 7.02 ± 1.211 |
8.285 ± 0.891 | 3.337 ± 0.761 |
2.186 ± 0.772 | 3.337 ± 0.811 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
