
CRESS virus sp. ctWOo3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; unclassified Cressdnaviricota; CRESS viruses
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
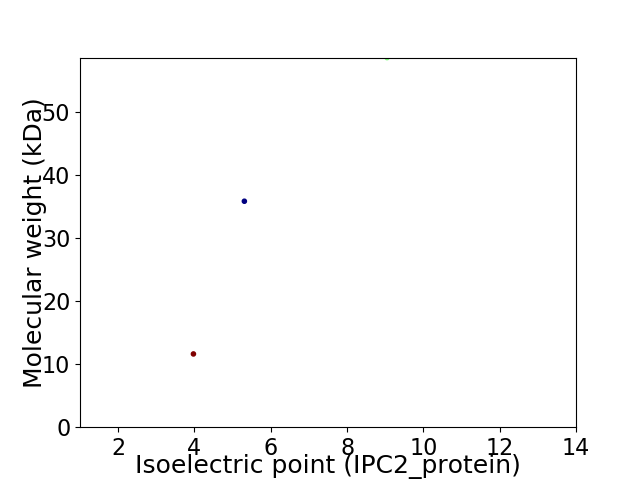
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
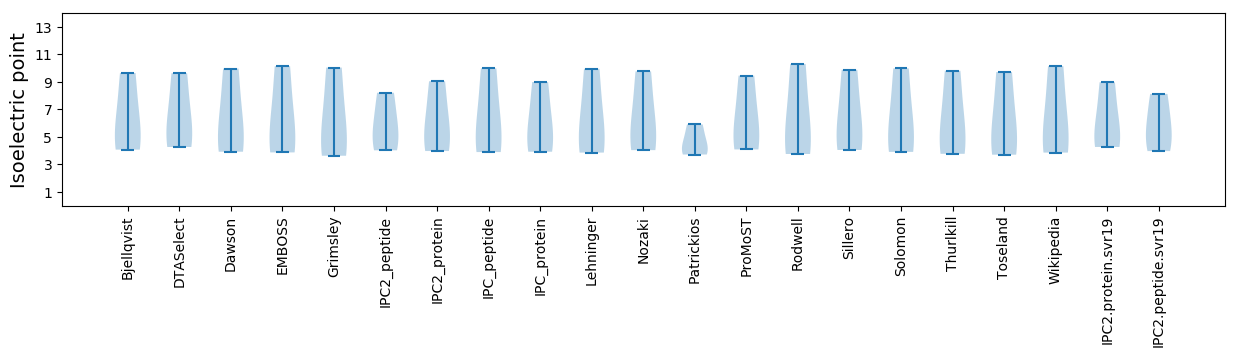
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2WC86|A0A5Q2WC86_9VIRU Putative capsid protein OS=CRESS virus sp. ctWOo3 OX=2656678 PE=4 SV=1
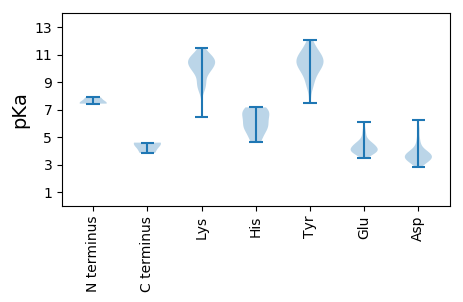
MM1 pKa = 7.88SDD3 pKa = 3.3RR4 pKa = 11.84LDD6 pKa = 3.46TVSISRR12 pKa = 11.84ACARR16 pKa = 11.84WLIDD20 pKa = 3.38YY21 pKa = 8.74GRR23 pKa = 11.84QLVNDD28 pKa = 5.65GYY30 pKa = 10.59GTDD33 pKa = 3.92YY34 pKa = 11.67DD35 pKa = 4.33PFNPVADD42 pKa = 5.05LEE44 pKa = 4.46PEE46 pKa = 4.15GAGFIDD52 pKa = 6.26DD53 pKa = 3.87IPMEE57 pKa = 4.55NGEE60 pKa = 3.99MWRR63 pKa = 11.84FMNALINFARR73 pKa = 11.84TVAGVLARR81 pKa = 11.84NMNDD85 pKa = 3.69PISPLTVSDD94 pKa = 5.45LFLEE98 pKa = 4.51DD99 pKa = 3.4EE100 pKa = 4.57KK101 pKa = 11.51EE102 pKa = 3.86II103 pKa = 4.59
MM1 pKa = 7.88SDD3 pKa = 3.3RR4 pKa = 11.84LDD6 pKa = 3.46TVSISRR12 pKa = 11.84ACARR16 pKa = 11.84WLIDD20 pKa = 3.38YY21 pKa = 8.74GRR23 pKa = 11.84QLVNDD28 pKa = 5.65GYY30 pKa = 10.59GTDD33 pKa = 3.92YY34 pKa = 11.67DD35 pKa = 4.33PFNPVADD42 pKa = 5.05LEE44 pKa = 4.46PEE46 pKa = 4.15GAGFIDD52 pKa = 6.26DD53 pKa = 3.87IPMEE57 pKa = 4.55NGEE60 pKa = 3.99MWRR63 pKa = 11.84FMNALINFARR73 pKa = 11.84TVAGVLARR81 pKa = 11.84NMNDD85 pKa = 3.69PISPLTVSDD94 pKa = 5.45LFLEE98 pKa = 4.51DD99 pKa = 3.4EE100 pKa = 4.57KK101 pKa = 11.51EE102 pKa = 3.86II103 pKa = 4.59
Molecular weight: 11.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2WC86|A0A5Q2WC86_9VIRU Putative capsid protein OS=CRESS virus sp. ctWOo3 OX=2656678 PE=4 SV=1
MM1 pKa = 7.43RR2 pKa = 11.84RR3 pKa = 11.84KK4 pKa = 10.42LFVSPFAKK12 pKa = 9.89PQGAVRR18 pKa = 11.84PNRR21 pKa = 11.84GQANKK26 pKa = 10.36KK27 pKa = 9.82SYY29 pKa = 9.57TYY31 pKa = 10.73RR32 pKa = 11.84KK33 pKa = 6.47NTRR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.87FSPGGFDD45 pKa = 3.19TDD47 pKa = 3.02KK48 pKa = 10.58MLRR51 pKa = 11.84NHH53 pKa = 7.01IDD55 pKa = 3.21NVTKK59 pKa = 10.64KK60 pKa = 10.7SIEE63 pKa = 4.11RR64 pKa = 11.84QKK66 pKa = 11.03KK67 pKa = 8.73PSFDD71 pKa = 3.35SVAPFVEE78 pKa = 4.7EE79 pKa = 4.01KK80 pKa = 10.13RR81 pKa = 11.84RR82 pKa = 11.84KK83 pKa = 9.21RR84 pKa = 11.84DD85 pKa = 3.48FKK87 pKa = 10.84IRR89 pKa = 11.84TSVGTSFHH97 pKa = 6.86VGPIPPPANGTRR109 pKa = 11.84QDD111 pKa = 4.41DD112 pKa = 3.96SSDD115 pKa = 3.55DD116 pKa = 3.09TKK118 pKa = 10.64MIHH121 pKa = 5.91KK122 pKa = 10.31VEE124 pKa = 4.25VKK126 pKa = 10.23SIVMGSDD133 pKa = 3.47PSQEE137 pKa = 3.68FRR139 pKa = 11.84RR140 pKa = 11.84KK141 pKa = 9.71IEE143 pKa = 3.94TGRR146 pKa = 11.84PTTKK150 pKa = 10.54SLLRR154 pKa = 11.84AAKK157 pKa = 10.34EE158 pKa = 3.75NGTTRR163 pKa = 11.84RR164 pKa = 11.84IIYY167 pKa = 9.79DD168 pKa = 3.72SYY170 pKa = 11.77NSAITDD176 pKa = 3.61GLHH179 pKa = 5.66FTRR182 pKa = 11.84KK183 pKa = 8.92QLTVGTGFNQRR194 pKa = 11.84GFTVFGVPAYY204 pKa = 7.45MTYY207 pKa = 11.31GDD209 pKa = 4.31MLGITDD215 pKa = 3.82TNLNNTQSAFKK226 pKa = 9.71MQTLLAGIMSSKK238 pKa = 10.04TEE240 pKa = 3.73MLLRR244 pKa = 11.84NQSAHH249 pKa = 4.88LRR251 pKa = 11.84AYY253 pKa = 10.32VRR255 pKa = 11.84VHH257 pKa = 6.58LVSKK261 pKa = 10.73QCGEE265 pKa = 4.22VNDD268 pKa = 4.82PVLDD272 pKa = 3.56MSNNVSHH279 pKa = 7.16GDD281 pKa = 3.48VNAEE285 pKa = 4.77SIPADD290 pKa = 3.88LKK292 pKa = 10.17NKK294 pKa = 9.74RR295 pKa = 11.84MPVTYY300 pKa = 10.12QYY302 pKa = 11.06TDD304 pKa = 2.81NRR306 pKa = 11.84LEE308 pKa = 4.19TGSTGTVEE316 pKa = 4.8LFPPSRR322 pKa = 11.84TLSFDD327 pKa = 3.09TTTKK331 pKa = 11.03GNILGQSPYY340 pKa = 10.26FKK342 pKa = 10.9EE343 pKa = 3.91NFKK346 pKa = 10.49ICATVSQNLGPGDD359 pKa = 4.06FFRR362 pKa = 11.84FSHH365 pKa = 4.66VHH367 pKa = 5.82HH368 pKa = 6.79YY369 pKa = 11.2GSGFDD374 pKa = 3.78LANLYY379 pKa = 9.58SEE381 pKa = 5.08KK382 pKa = 10.84SLTGPDD388 pKa = 3.39DD389 pKa = 4.06PGYY392 pKa = 11.11AKK394 pKa = 8.32EE395 pKa = 3.86TSYY398 pKa = 11.12FYY400 pKa = 9.86IVEE403 pKa = 4.39YY404 pKa = 10.21KK405 pKa = 10.91GHH407 pKa = 5.89ICEE410 pKa = 4.33ATFNNAGAFEE420 pKa = 4.16ARR422 pKa = 11.84IGTAPVLLNAEE433 pKa = 3.87IRR435 pKa = 11.84KK436 pKa = 9.11SVRR439 pKa = 11.84FVNAPNSGQDD449 pKa = 3.21QTNSGVAPRR458 pKa = 11.84CLLRR462 pKa = 11.84VFEE465 pKa = 4.58QSSLPNIVEE474 pKa = 3.69QRR476 pKa = 11.84RR477 pKa = 11.84DD478 pKa = 3.69FNSDD482 pKa = 2.9LTNWTTVPTPALGLYY497 pKa = 9.19VIQLSSDD504 pKa = 3.06RR505 pKa = 11.84TLRR508 pKa = 11.84TEE510 pKa = 3.96VNQAGNSNATDD521 pKa = 3.45IVPP524 pKa = 3.84
MM1 pKa = 7.43RR2 pKa = 11.84RR3 pKa = 11.84KK4 pKa = 10.42LFVSPFAKK12 pKa = 9.89PQGAVRR18 pKa = 11.84PNRR21 pKa = 11.84GQANKK26 pKa = 10.36KK27 pKa = 9.82SYY29 pKa = 9.57TYY31 pKa = 10.73RR32 pKa = 11.84KK33 pKa = 6.47NTRR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.87FSPGGFDD45 pKa = 3.19TDD47 pKa = 3.02KK48 pKa = 10.58MLRR51 pKa = 11.84NHH53 pKa = 7.01IDD55 pKa = 3.21NVTKK59 pKa = 10.64KK60 pKa = 10.7SIEE63 pKa = 4.11RR64 pKa = 11.84QKK66 pKa = 11.03KK67 pKa = 8.73PSFDD71 pKa = 3.35SVAPFVEE78 pKa = 4.7EE79 pKa = 4.01KK80 pKa = 10.13RR81 pKa = 11.84RR82 pKa = 11.84KK83 pKa = 9.21RR84 pKa = 11.84DD85 pKa = 3.48FKK87 pKa = 10.84IRR89 pKa = 11.84TSVGTSFHH97 pKa = 6.86VGPIPPPANGTRR109 pKa = 11.84QDD111 pKa = 4.41DD112 pKa = 3.96SSDD115 pKa = 3.55DD116 pKa = 3.09TKK118 pKa = 10.64MIHH121 pKa = 5.91KK122 pKa = 10.31VEE124 pKa = 4.25VKK126 pKa = 10.23SIVMGSDD133 pKa = 3.47PSQEE137 pKa = 3.68FRR139 pKa = 11.84RR140 pKa = 11.84KK141 pKa = 9.71IEE143 pKa = 3.94TGRR146 pKa = 11.84PTTKK150 pKa = 10.54SLLRR154 pKa = 11.84AAKK157 pKa = 10.34EE158 pKa = 3.75NGTTRR163 pKa = 11.84RR164 pKa = 11.84IIYY167 pKa = 9.79DD168 pKa = 3.72SYY170 pKa = 11.77NSAITDD176 pKa = 3.61GLHH179 pKa = 5.66FTRR182 pKa = 11.84KK183 pKa = 8.92QLTVGTGFNQRR194 pKa = 11.84GFTVFGVPAYY204 pKa = 7.45MTYY207 pKa = 11.31GDD209 pKa = 4.31MLGITDD215 pKa = 3.82TNLNNTQSAFKK226 pKa = 9.71MQTLLAGIMSSKK238 pKa = 10.04TEE240 pKa = 3.73MLLRR244 pKa = 11.84NQSAHH249 pKa = 4.88LRR251 pKa = 11.84AYY253 pKa = 10.32VRR255 pKa = 11.84VHH257 pKa = 6.58LVSKK261 pKa = 10.73QCGEE265 pKa = 4.22VNDD268 pKa = 4.82PVLDD272 pKa = 3.56MSNNVSHH279 pKa = 7.16GDD281 pKa = 3.48VNAEE285 pKa = 4.77SIPADD290 pKa = 3.88LKK292 pKa = 10.17NKK294 pKa = 9.74RR295 pKa = 11.84MPVTYY300 pKa = 10.12QYY302 pKa = 11.06TDD304 pKa = 2.81NRR306 pKa = 11.84LEE308 pKa = 4.19TGSTGTVEE316 pKa = 4.8LFPPSRR322 pKa = 11.84TLSFDD327 pKa = 3.09TTTKK331 pKa = 11.03GNILGQSPYY340 pKa = 10.26FKK342 pKa = 10.9EE343 pKa = 3.91NFKK346 pKa = 10.49ICATVSQNLGPGDD359 pKa = 4.06FFRR362 pKa = 11.84FSHH365 pKa = 4.66VHH367 pKa = 5.82HH368 pKa = 6.79YY369 pKa = 11.2GSGFDD374 pKa = 3.78LANLYY379 pKa = 9.58SEE381 pKa = 5.08KK382 pKa = 10.84SLTGPDD388 pKa = 3.39DD389 pKa = 4.06PGYY392 pKa = 11.11AKK394 pKa = 8.32EE395 pKa = 3.86TSYY398 pKa = 11.12FYY400 pKa = 9.86IVEE403 pKa = 4.39YY404 pKa = 10.21KK405 pKa = 10.91GHH407 pKa = 5.89ICEE410 pKa = 4.33ATFNNAGAFEE420 pKa = 4.16ARR422 pKa = 11.84IGTAPVLLNAEE433 pKa = 3.87IRR435 pKa = 11.84KK436 pKa = 9.11SVRR439 pKa = 11.84FVNAPNSGQDD449 pKa = 3.21QTNSGVAPRR458 pKa = 11.84CLLRR462 pKa = 11.84VFEE465 pKa = 4.58QSSLPNIVEE474 pKa = 3.69QRR476 pKa = 11.84RR477 pKa = 11.84DD478 pKa = 3.69FNSDD482 pKa = 2.9LTNWTTVPTPALGLYY497 pKa = 9.19VIQLSSDD504 pKa = 3.06RR505 pKa = 11.84TLRR508 pKa = 11.84TEE510 pKa = 3.96VNQAGNSNATDD521 pKa = 3.45IVPP524 pKa = 3.84
Molecular weight: 58.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
932 |
103 |
524 |
310.7 |
35.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.721 ± 1.184 | 0.966 ± 0.164 |
7.296 ± 1.582 | 5.043 ± 0.719 |
5.687 ± 0.402 | 6.116 ± 0.837 |
1.395 ± 0.592 | 5.687 ± 1.164 |
6.223 ± 1.396 | 7.618 ± 0.885 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.682 ± 0.644 | 6.438 ± 0.241 |
5.043 ± 0.5 | 3.219 ± 0.664 |
6.545 ± 0.597 | 7.403 ± 0.807 |
6.974 ± 1.475 | 6.438 ± 0.219 |
0.966 ± 0.613 | 3.541 ± 0.33 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
