
Sewage-associated circular DNA virus-7
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.59
Get precalculated fractions of proteins
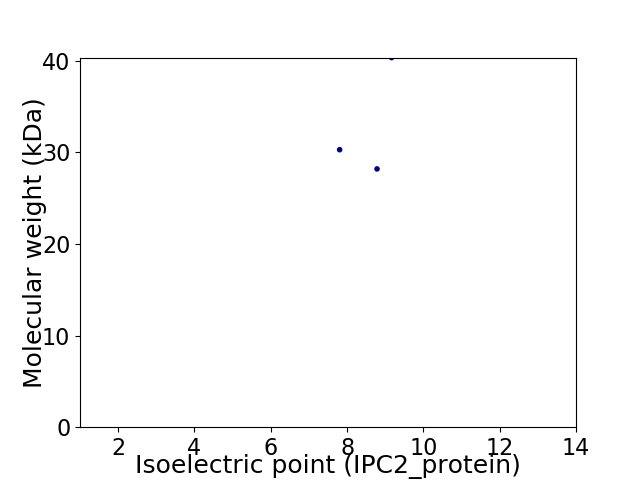
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
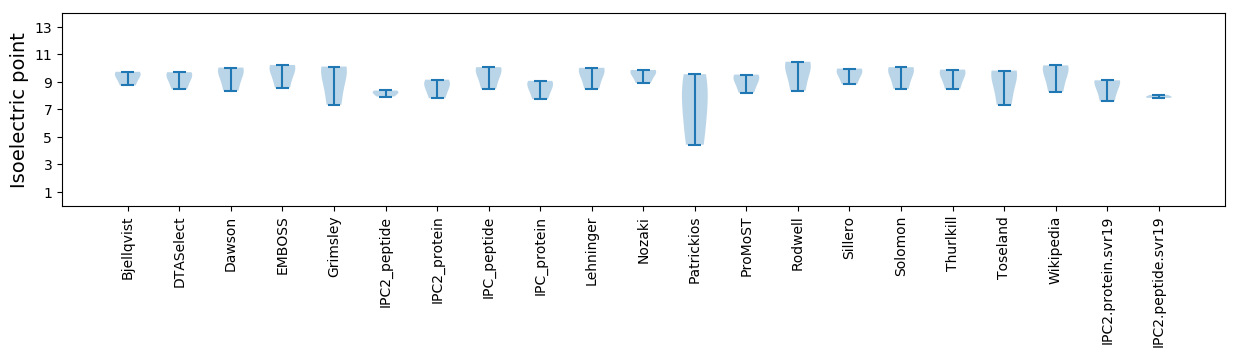
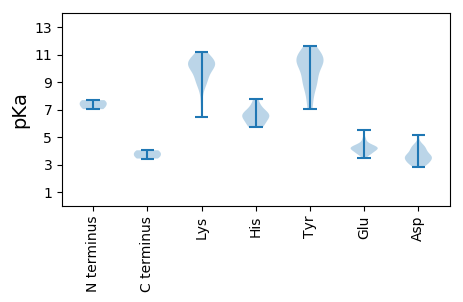
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075J468|A0A075J468_9VIRU Putative capsid protein OS=Sewage-associated circular DNA virus-7 OX=1519396 PE=4 SV=1
MM1 pKa = 7.68ARR3 pKa = 11.84RR4 pKa = 11.84QGIYY8 pKa = 9.05WLLTIPHH15 pKa = 6.26SEE17 pKa = 4.05FTPYY21 pKa = 10.64LPPPVAWIRR30 pKa = 11.84GQLEE34 pKa = 4.24LGAGGYY40 pKa = 7.29LHH42 pKa = 6.69WQAVLAFRR50 pKa = 11.84SKK52 pKa = 11.21GSLATVQAIFGTAMHH67 pKa = 6.82AEE69 pKa = 4.42ISRR72 pKa = 11.84SSAASDD78 pKa = 3.91YY79 pKa = 10.57VWKK82 pKa = 10.46EE83 pKa = 3.8DD84 pKa = 3.46TRR86 pKa = 11.84VSGTQFEE93 pKa = 4.53LGEE96 pKa = 4.27RR97 pKa = 11.84PFKK100 pKa = 11.15RR101 pKa = 11.84NDD103 pKa = 3.05PRR105 pKa = 11.84DD106 pKa = 3.12WDD108 pKa = 5.17AIWEE112 pKa = 4.2MAVAGNLCGIPASVRR127 pKa = 11.84VQSYY131 pKa = 7.02RR132 pKa = 11.84TLRR135 pKa = 11.84AICADD140 pKa = 3.05HH141 pKa = 6.49SKK143 pKa = 10.23PVGMEE148 pKa = 3.66RR149 pKa = 11.84TCFVFWGRR157 pKa = 11.84TGTGKK162 pKa = 9.99SRR164 pKa = 11.84RR165 pKa = 11.84AWDD168 pKa = 3.43EE169 pKa = 4.06AGVDD173 pKa = 4.36AYY175 pKa = 11.56SKK177 pKa = 11.13DD178 pKa = 3.53PNTKK182 pKa = 9.54FWCGYY187 pKa = 7.95NGQRR191 pKa = 11.84NVVIDD196 pKa = 3.81EE197 pKa = 3.85FRR199 pKa = 11.84GRR201 pKa = 11.84IDD203 pKa = 3.04VSHH206 pKa = 7.15FLRR209 pKa = 11.84WLDD212 pKa = 3.54RR213 pKa = 11.84YY214 pKa = 9.27PVNVEE219 pKa = 3.75IKK221 pKa = 10.51GSSTPLCCEE230 pKa = 3.95RR231 pKa = 11.84VWITSNLDD239 pKa = 2.94PRR241 pKa = 11.84AWYY244 pKa = 10.44SEE246 pKa = 3.88LDD248 pKa = 3.84EE249 pKa = 4.47EE250 pKa = 4.85TLNALLRR257 pKa = 11.84RR258 pKa = 11.84LNITHH263 pKa = 7.31FLL265 pKa = 3.74
MM1 pKa = 7.68ARR3 pKa = 11.84RR4 pKa = 11.84QGIYY8 pKa = 9.05WLLTIPHH15 pKa = 6.26SEE17 pKa = 4.05FTPYY21 pKa = 10.64LPPPVAWIRR30 pKa = 11.84GQLEE34 pKa = 4.24LGAGGYY40 pKa = 7.29LHH42 pKa = 6.69WQAVLAFRR50 pKa = 11.84SKK52 pKa = 11.21GSLATVQAIFGTAMHH67 pKa = 6.82AEE69 pKa = 4.42ISRR72 pKa = 11.84SSAASDD78 pKa = 3.91YY79 pKa = 10.57VWKK82 pKa = 10.46EE83 pKa = 3.8DD84 pKa = 3.46TRR86 pKa = 11.84VSGTQFEE93 pKa = 4.53LGEE96 pKa = 4.27RR97 pKa = 11.84PFKK100 pKa = 11.15RR101 pKa = 11.84NDD103 pKa = 3.05PRR105 pKa = 11.84DD106 pKa = 3.12WDD108 pKa = 5.17AIWEE112 pKa = 4.2MAVAGNLCGIPASVRR127 pKa = 11.84VQSYY131 pKa = 7.02RR132 pKa = 11.84TLRR135 pKa = 11.84AICADD140 pKa = 3.05HH141 pKa = 6.49SKK143 pKa = 10.23PVGMEE148 pKa = 3.66RR149 pKa = 11.84TCFVFWGRR157 pKa = 11.84TGTGKK162 pKa = 9.99SRR164 pKa = 11.84RR165 pKa = 11.84AWDD168 pKa = 3.43EE169 pKa = 4.06AGVDD173 pKa = 4.36AYY175 pKa = 11.56SKK177 pKa = 11.13DD178 pKa = 3.53PNTKK182 pKa = 9.54FWCGYY187 pKa = 7.95NGQRR191 pKa = 11.84NVVIDD196 pKa = 3.81EE197 pKa = 3.85FRR199 pKa = 11.84GRR201 pKa = 11.84IDD203 pKa = 3.04VSHH206 pKa = 7.15FLRR209 pKa = 11.84WLDD212 pKa = 3.54RR213 pKa = 11.84YY214 pKa = 9.27PVNVEE219 pKa = 3.75IKK221 pKa = 10.51GSSTPLCCEE230 pKa = 3.95RR231 pKa = 11.84VWITSNLDD239 pKa = 2.94PRR241 pKa = 11.84AWYY244 pKa = 10.44SEE246 pKa = 3.88LDD248 pKa = 3.84EE249 pKa = 4.47EE250 pKa = 4.85TLNALLRR257 pKa = 11.84RR258 pKa = 11.84LNITHH263 pKa = 7.31FLL265 pKa = 3.74
Molecular weight: 30.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075J468|A0A075J468_9VIRU Putative capsid protein OS=Sewage-associated circular DNA virus-7 OX=1519396 PE=4 SV=1
MM1 pKa = 7.39IKK3 pKa = 10.07KK4 pKa = 9.51HH5 pKa = 5.81FKK7 pKa = 10.27SIKK10 pKa = 9.78KK11 pKa = 10.14GIAAAATAASLRR23 pKa = 11.84RR24 pKa = 11.84SNQQRR29 pKa = 11.84LAQRR33 pKa = 11.84KK34 pKa = 8.09RR35 pKa = 11.84VGRR38 pKa = 11.84SRR40 pKa = 11.84TKK42 pKa = 10.38TKK44 pKa = 10.28TKK46 pKa = 8.93TKK48 pKa = 10.04RR49 pKa = 11.84FRR51 pKa = 11.84TITDD55 pKa = 3.17DD56 pKa = 3.68SGIGSSAHH64 pKa = 5.74LVVVDD69 pKa = 4.23AKK71 pKa = 10.71KK72 pKa = 10.8KK73 pKa = 9.04KK74 pKa = 6.44TAKK77 pKa = 10.33GKK79 pKa = 8.89WIYY82 pKa = 8.8TQTHH86 pKa = 5.72NEE88 pKa = 4.29VIFSNAGTQGTQQLGMGALSQINVSSGVGYY118 pKa = 8.88TGFQSQVAVRR128 pKa = 11.84QLNPMLGNTGSAYY141 pKa = 9.3LTASTAPTTDD151 pKa = 2.84SFVISDD157 pKa = 4.18MIFDD161 pKa = 4.51IEE163 pKa = 4.22MCSWSDD169 pKa = 3.4VAQTVDD175 pKa = 3.17MYY177 pKa = 11.63VYY179 pKa = 10.11VAKK182 pKa = 10.61DD183 pKa = 3.32GQANDD188 pKa = 4.33VGIEE192 pKa = 3.85WARR195 pKa = 11.84GYY197 pKa = 8.37QQQGSGKK204 pKa = 10.26GAMTMLSAPLYY215 pKa = 10.1QGVAGYY221 pKa = 10.96GRR223 pKa = 11.84VDD225 pKa = 3.56EE226 pKa = 5.49PYY228 pKa = 11.03ASPLDD233 pKa = 3.61VKK235 pKa = 10.92EE236 pKa = 4.12YY237 pKa = 10.62MNNTYY242 pKa = 10.84RR243 pKa = 11.84FMKK246 pKa = 10.25KK247 pKa = 7.39VTINLTSGATHH258 pKa = 6.72KK259 pKa = 10.96CKK261 pKa = 10.47VHH263 pKa = 6.72CVMNKK268 pKa = 9.69LVNNSEE274 pKa = 4.33LDD276 pKa = 3.27VDD278 pKa = 4.14TRR280 pKa = 11.84GSDD283 pKa = 3.48LTIKK287 pKa = 10.71GLTTIIMITVRR298 pKa = 11.84GQVVDD303 pKa = 3.59DD304 pKa = 3.94TTAGGTAKK312 pKa = 9.21PTFASTRR319 pKa = 11.84VGLITRR325 pKa = 11.84NTYY328 pKa = 9.22VCHH331 pKa = 6.27PVPSPISNRR340 pKa = 11.84TVGIHH345 pKa = 6.43SYY347 pKa = 10.44NVPINAALAAQQLINDD363 pKa = 3.98VDD365 pKa = 4.29VVAQAINCC373 pKa = 4.04
MM1 pKa = 7.39IKK3 pKa = 10.07KK4 pKa = 9.51HH5 pKa = 5.81FKK7 pKa = 10.27SIKK10 pKa = 9.78KK11 pKa = 10.14GIAAAATAASLRR23 pKa = 11.84RR24 pKa = 11.84SNQQRR29 pKa = 11.84LAQRR33 pKa = 11.84KK34 pKa = 8.09RR35 pKa = 11.84VGRR38 pKa = 11.84SRR40 pKa = 11.84TKK42 pKa = 10.38TKK44 pKa = 10.28TKK46 pKa = 8.93TKK48 pKa = 10.04RR49 pKa = 11.84FRR51 pKa = 11.84TITDD55 pKa = 3.17DD56 pKa = 3.68SGIGSSAHH64 pKa = 5.74LVVVDD69 pKa = 4.23AKK71 pKa = 10.71KK72 pKa = 10.8KK73 pKa = 9.04KK74 pKa = 6.44TAKK77 pKa = 10.33GKK79 pKa = 8.89WIYY82 pKa = 8.8TQTHH86 pKa = 5.72NEE88 pKa = 4.29VIFSNAGTQGTQQLGMGALSQINVSSGVGYY118 pKa = 8.88TGFQSQVAVRR128 pKa = 11.84QLNPMLGNTGSAYY141 pKa = 9.3LTASTAPTTDD151 pKa = 2.84SFVISDD157 pKa = 4.18MIFDD161 pKa = 4.51IEE163 pKa = 4.22MCSWSDD169 pKa = 3.4VAQTVDD175 pKa = 3.17MYY177 pKa = 11.63VYY179 pKa = 10.11VAKK182 pKa = 10.61DD183 pKa = 3.32GQANDD188 pKa = 4.33VGIEE192 pKa = 3.85WARR195 pKa = 11.84GYY197 pKa = 8.37QQQGSGKK204 pKa = 10.26GAMTMLSAPLYY215 pKa = 10.1QGVAGYY221 pKa = 10.96GRR223 pKa = 11.84VDD225 pKa = 3.56EE226 pKa = 5.49PYY228 pKa = 11.03ASPLDD233 pKa = 3.61VKK235 pKa = 10.92EE236 pKa = 4.12YY237 pKa = 10.62MNNTYY242 pKa = 10.84RR243 pKa = 11.84FMKK246 pKa = 10.25KK247 pKa = 7.39VTINLTSGATHH258 pKa = 6.72KK259 pKa = 10.96CKK261 pKa = 10.47VHH263 pKa = 6.72CVMNKK268 pKa = 9.69LVNNSEE274 pKa = 4.33LDD276 pKa = 3.27VDD278 pKa = 4.14TRR280 pKa = 11.84GSDD283 pKa = 3.48LTIKK287 pKa = 10.71GLTTIIMITVRR298 pKa = 11.84GQVVDD303 pKa = 3.59DD304 pKa = 3.94TTAGGTAKK312 pKa = 9.21PTFASTRR319 pKa = 11.84VGLITRR325 pKa = 11.84NTYY328 pKa = 9.22VCHH331 pKa = 6.27PVPSPISNRR340 pKa = 11.84TVGIHH345 pKa = 6.43SYY347 pKa = 10.44NVPINAALAAQQLINDD363 pKa = 3.98VDD365 pKa = 4.29VVAQAINCC373 pKa = 4.04
Molecular weight: 40.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
898 |
260 |
373 |
299.3 |
32.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.572 ± 1.0 | 1.559 ± 0.251 |
4.009 ± 0.921 | 4.12 ± 1.257 |
2.784 ± 0.503 | 7.795 ± 0.259 |
1.782 ± 0.215 | 5.122 ± 0.544 |
5.011 ± 1.034 | 7.35 ± 1.165 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.561 ± 0.419 | 3.898 ± 0.603 |
5.679 ± 1.794 | 4.009 ± 0.827 |
6.459 ± 1.136 | 9.688 ± 1.959 |
8.018 ± 1.069 | 7.238 ± 0.945 |
2.45 ± 0.932 | 2.895 ± 0.443 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
