
Avon-Heathcote Estuary associated circular virus 26
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.65
Get precalculated fractions of proteins
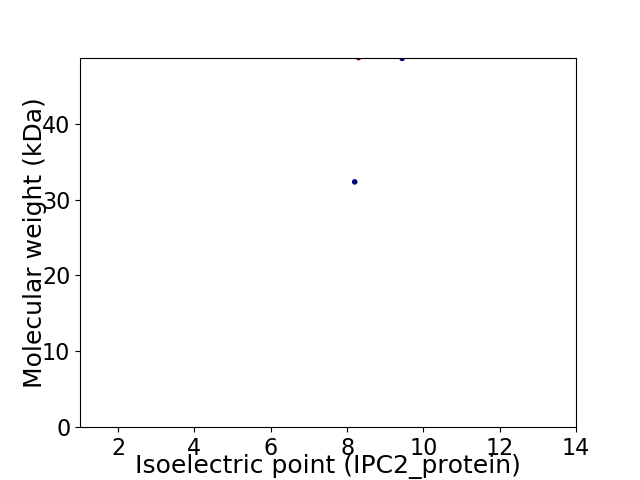
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IB96|A0A0C5IB96_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 26 OX=1618250 PE=4 SV=1
MM1 pKa = 8.04PSRR4 pKa = 11.84QGVYY8 pKa = 9.87FIITVPYY15 pKa = 9.5EE16 pKa = 4.15KK17 pKa = 10.65YY18 pKa = 9.72EE19 pKa = 4.2KK20 pKa = 10.24PIEE23 pKa = 4.02LPRR26 pKa = 11.84GFRR29 pKa = 11.84YY30 pKa = 9.86ISGQGEE36 pKa = 4.05IGASGYY42 pKa = 8.92HH43 pKa = 5.97HH44 pKa = 6.61WQIICAFEE52 pKa = 3.96QKK54 pKa = 8.91KK55 pKa = 7.49TLTGAKK61 pKa = 9.84SYY63 pKa = 8.62FAPEE67 pKa = 3.38AHH69 pKa = 7.22LEE71 pKa = 4.2FTRR74 pKa = 11.84SRR76 pKa = 11.84CARR79 pKa = 11.84DD80 pKa = 3.41YY81 pKa = 10.76CHH83 pKa = 7.17KK84 pKa = 10.45EE85 pKa = 3.62EE86 pKa = 4.15TRR88 pKa = 11.84IPDD91 pKa = 3.11SSFEE95 pKa = 4.03FGTYY99 pKa = 6.87PTRR102 pKa = 11.84VNNKK106 pKa = 8.8QDD108 pKa = 3.0WDD110 pKa = 3.99LVLEE114 pKa = 4.31KK115 pKa = 10.64AKK117 pKa = 10.54KK118 pKa = 10.06GQFDD122 pKa = 5.51LIDD125 pKa = 4.46GDD127 pKa = 3.95VMMRR131 pKa = 11.84HH132 pKa = 5.95YY133 pKa = 11.66NNIRR137 pKa = 11.84KK138 pKa = 9.42IYY140 pKa = 10.54SDD142 pKa = 3.27FQKK145 pKa = 10.25PVKK148 pKa = 10.53RR149 pKa = 11.84EE150 pKa = 3.81VQVVNVYY157 pKa = 8.78WGQTGTGKK165 pKa = 8.65TKK167 pKa = 10.75SVFEE171 pKa = 4.49EE172 pKa = 4.12IGDD175 pKa = 3.79DD176 pKa = 4.4FYY178 pKa = 12.2VKK180 pKa = 10.07MPSTKK185 pKa = 9.74WFDD188 pKa = 3.49GYY190 pKa = 10.66RR191 pKa = 11.84GQEE194 pKa = 3.57NVVIDD199 pKa = 4.35EE200 pKa = 4.3FTGQVDD206 pKa = 3.39ITHH209 pKa = 7.62LLRR212 pKa = 11.84WLDD215 pKa = 3.62QYY217 pKa = 11.47PCTVEE222 pKa = 4.5VKK224 pKa = 10.54GSQVFLNTKK233 pKa = 9.37KK234 pKa = 9.22WWITSNIDD242 pKa = 3.1PAKK245 pKa = 9.17WYY247 pKa = 9.39EE248 pKa = 4.23GKK250 pKa = 10.33DD251 pKa = 3.41EE252 pKa = 4.24EE253 pKa = 5.19QIKK256 pKa = 9.95ALKK259 pKa = 10.24RR260 pKa = 11.84RR261 pKa = 11.84FTNVVHH267 pKa = 5.75YY268 pKa = 8.68LKK270 pKa = 10.42PFKK273 pKa = 10.51KK274 pKa = 10.45
MM1 pKa = 8.04PSRR4 pKa = 11.84QGVYY8 pKa = 9.87FIITVPYY15 pKa = 9.5EE16 pKa = 4.15KK17 pKa = 10.65YY18 pKa = 9.72EE19 pKa = 4.2KK20 pKa = 10.24PIEE23 pKa = 4.02LPRR26 pKa = 11.84GFRR29 pKa = 11.84YY30 pKa = 9.86ISGQGEE36 pKa = 4.05IGASGYY42 pKa = 8.92HH43 pKa = 5.97HH44 pKa = 6.61WQIICAFEE52 pKa = 3.96QKK54 pKa = 8.91KK55 pKa = 7.49TLTGAKK61 pKa = 9.84SYY63 pKa = 8.62FAPEE67 pKa = 3.38AHH69 pKa = 7.22LEE71 pKa = 4.2FTRR74 pKa = 11.84SRR76 pKa = 11.84CARR79 pKa = 11.84DD80 pKa = 3.41YY81 pKa = 10.76CHH83 pKa = 7.17KK84 pKa = 10.45EE85 pKa = 3.62EE86 pKa = 4.15TRR88 pKa = 11.84IPDD91 pKa = 3.11SSFEE95 pKa = 4.03FGTYY99 pKa = 6.87PTRR102 pKa = 11.84VNNKK106 pKa = 8.8QDD108 pKa = 3.0WDD110 pKa = 3.99LVLEE114 pKa = 4.31KK115 pKa = 10.64AKK117 pKa = 10.54KK118 pKa = 10.06GQFDD122 pKa = 5.51LIDD125 pKa = 4.46GDD127 pKa = 3.95VMMRR131 pKa = 11.84HH132 pKa = 5.95YY133 pKa = 11.66NNIRR137 pKa = 11.84KK138 pKa = 9.42IYY140 pKa = 10.54SDD142 pKa = 3.27FQKK145 pKa = 10.25PVKK148 pKa = 10.53RR149 pKa = 11.84EE150 pKa = 3.81VQVVNVYY157 pKa = 8.78WGQTGTGKK165 pKa = 8.65TKK167 pKa = 10.75SVFEE171 pKa = 4.49EE172 pKa = 4.12IGDD175 pKa = 3.79DD176 pKa = 4.4FYY178 pKa = 12.2VKK180 pKa = 10.07MPSTKK185 pKa = 9.74WFDD188 pKa = 3.49GYY190 pKa = 10.66RR191 pKa = 11.84GQEE194 pKa = 3.57NVVIDD199 pKa = 4.35EE200 pKa = 4.3FTGQVDD206 pKa = 3.39ITHH209 pKa = 7.62LLRR212 pKa = 11.84WLDD215 pKa = 3.62QYY217 pKa = 11.47PCTVEE222 pKa = 4.5VKK224 pKa = 10.54GSQVFLNTKK233 pKa = 9.37KK234 pKa = 9.22WWITSNIDD242 pKa = 3.1PAKK245 pKa = 9.17WYY247 pKa = 9.39EE248 pKa = 4.23GKK250 pKa = 10.33DD251 pKa = 3.41EE252 pKa = 4.24EE253 pKa = 5.19QIKK256 pKa = 9.95ALKK259 pKa = 10.24RR260 pKa = 11.84RR261 pKa = 11.84FTNVVHH267 pKa = 5.75YY268 pKa = 8.68LKK270 pKa = 10.42PFKK273 pKa = 10.51KK274 pKa = 10.45
Molecular weight: 32.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IB96|A0A0C5IB96_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 26 OX=1618250 PE=4 SV=1
MM1 pKa = 8.14DD2 pKa = 3.68KK3 pKa = 10.66QIYY6 pKa = 10.55DD7 pKa = 3.45MAAKK11 pKa = 9.97YY12 pKa = 10.0IKK14 pKa = 10.52SNLGVHH20 pKa = 5.42VPSYY24 pKa = 8.69NTTRR28 pKa = 11.84TVYY31 pKa = 8.81NTYY34 pKa = 10.51RR35 pKa = 11.84NARR38 pKa = 11.84RR39 pKa = 11.84SMKK42 pKa = 9.49STNQKK47 pKa = 9.06MKK49 pKa = 10.67SSSSKK54 pKa = 8.61NWKK57 pKa = 8.3VSPRR61 pKa = 11.84ITYY64 pKa = 6.58PTSMSSTKK72 pKa = 10.0TAVLPPKK79 pKa = 10.37KK80 pKa = 9.78KK81 pKa = 9.15FRR83 pKa = 11.84KK84 pKa = 8.55IRR86 pKa = 11.84RR87 pKa = 11.84TIVAGSRR94 pKa = 11.84KK95 pKa = 9.36SVPKK99 pKa = 10.3KK100 pKa = 8.16VRR102 pKa = 11.84KK103 pKa = 8.79YY104 pKa = 10.39VKK106 pKa = 10.18KK107 pKa = 10.63AVNDD111 pKa = 3.55KK112 pKa = 9.98TKK114 pKa = 9.26YY115 pKa = 7.31TQGPGARR122 pKa = 11.84IEE124 pKa = 4.52LYY126 pKa = 9.31NTFHH130 pKa = 7.55LDD132 pKa = 2.88TGNSVIQTGQFGSSPGDD149 pKa = 3.58SNHH152 pKa = 6.35AFIPFFTDD160 pKa = 2.66TVMNTEE166 pKa = 3.98FRR168 pKa = 11.84IGNNPVGDD176 pKa = 4.15DD177 pKa = 4.45AGMLDD182 pKa = 4.58PPPAFNASSGASVQYY197 pKa = 10.77IKK199 pKa = 10.93DD200 pKa = 3.63NEE202 pKa = 4.79SIVIDD207 pKa = 4.15HH208 pKa = 6.32YY209 pKa = 11.54SATFFMTNTSDD220 pKa = 3.37TKK222 pKa = 11.6AMFHH226 pKa = 5.41IQEE229 pKa = 4.37WVCNRR234 pKa = 11.84DD235 pKa = 2.75SDD237 pKa = 4.24VNIVVRR243 pKa = 11.84TTEE246 pKa = 3.71QFNNQTWRR254 pKa = 11.84NDD256 pKa = 3.38PGFATIVPVSSTNLFKK272 pKa = 11.01QPNFEE277 pKa = 3.88ISKK280 pKa = 10.3VPGISQYY287 pKa = 8.42WKK289 pKa = 9.24KK290 pKa = 11.2GSIKK294 pKa = 10.31RR295 pKa = 11.84QFEE298 pKa = 4.38LMPGEE303 pKa = 4.6SIEE306 pKa = 4.52LSLPFGKK313 pKa = 10.33KK314 pKa = 7.52YY315 pKa = 10.28WNLQRR320 pKa = 11.84FNDD323 pKa = 3.75QNEE326 pKa = 4.34TLTGRR331 pKa = 11.84FEE333 pKa = 4.0YY334 pKa = 10.45HH335 pKa = 6.68KK336 pKa = 10.77NLSRR340 pKa = 11.84YY341 pKa = 8.62LQVSVRR347 pKa = 11.84GSVGFSADD355 pKa = 3.84TPPKK359 pKa = 10.65SSFQGTNVYY368 pKa = 10.7YY369 pKa = 10.55KK370 pKa = 10.55VLKK373 pKa = 9.98TMIAHH378 pKa = 6.14RR379 pKa = 11.84VDD381 pKa = 4.75RR382 pKa = 11.84FGQQPKK388 pKa = 9.74RR389 pKa = 11.84YY390 pKa = 8.84RR391 pKa = 11.84IIGNPSEE398 pKa = 4.43VVSSFNGTAVDD409 pKa = 4.36LGRR412 pKa = 11.84PAITEE417 pKa = 3.95PVEE420 pKa = 5.34SIIQTLQASTT430 pKa = 3.59
MM1 pKa = 8.14DD2 pKa = 3.68KK3 pKa = 10.66QIYY6 pKa = 10.55DD7 pKa = 3.45MAAKK11 pKa = 9.97YY12 pKa = 10.0IKK14 pKa = 10.52SNLGVHH20 pKa = 5.42VPSYY24 pKa = 8.69NTTRR28 pKa = 11.84TVYY31 pKa = 8.81NTYY34 pKa = 10.51RR35 pKa = 11.84NARR38 pKa = 11.84RR39 pKa = 11.84SMKK42 pKa = 9.49STNQKK47 pKa = 9.06MKK49 pKa = 10.67SSSSKK54 pKa = 8.61NWKK57 pKa = 8.3VSPRR61 pKa = 11.84ITYY64 pKa = 6.58PTSMSSTKK72 pKa = 10.0TAVLPPKK79 pKa = 10.37KK80 pKa = 9.78KK81 pKa = 9.15FRR83 pKa = 11.84KK84 pKa = 8.55IRR86 pKa = 11.84RR87 pKa = 11.84TIVAGSRR94 pKa = 11.84KK95 pKa = 9.36SVPKK99 pKa = 10.3KK100 pKa = 8.16VRR102 pKa = 11.84KK103 pKa = 8.79YY104 pKa = 10.39VKK106 pKa = 10.18KK107 pKa = 10.63AVNDD111 pKa = 3.55KK112 pKa = 9.98TKK114 pKa = 9.26YY115 pKa = 7.31TQGPGARR122 pKa = 11.84IEE124 pKa = 4.52LYY126 pKa = 9.31NTFHH130 pKa = 7.55LDD132 pKa = 2.88TGNSVIQTGQFGSSPGDD149 pKa = 3.58SNHH152 pKa = 6.35AFIPFFTDD160 pKa = 2.66TVMNTEE166 pKa = 3.98FRR168 pKa = 11.84IGNNPVGDD176 pKa = 4.15DD177 pKa = 4.45AGMLDD182 pKa = 4.58PPPAFNASSGASVQYY197 pKa = 10.77IKK199 pKa = 10.93DD200 pKa = 3.63NEE202 pKa = 4.79SIVIDD207 pKa = 4.15HH208 pKa = 6.32YY209 pKa = 11.54SATFFMTNTSDD220 pKa = 3.37TKK222 pKa = 11.6AMFHH226 pKa = 5.41IQEE229 pKa = 4.37WVCNRR234 pKa = 11.84DD235 pKa = 2.75SDD237 pKa = 4.24VNIVVRR243 pKa = 11.84TTEE246 pKa = 3.71QFNNQTWRR254 pKa = 11.84NDD256 pKa = 3.38PGFATIVPVSSTNLFKK272 pKa = 11.01QPNFEE277 pKa = 3.88ISKK280 pKa = 10.3VPGISQYY287 pKa = 8.42WKK289 pKa = 9.24KK290 pKa = 11.2GSIKK294 pKa = 10.31RR295 pKa = 11.84QFEE298 pKa = 4.38LMPGEE303 pKa = 4.6SIEE306 pKa = 4.52LSLPFGKK313 pKa = 10.33KK314 pKa = 7.52YY315 pKa = 10.28WNLQRR320 pKa = 11.84FNDD323 pKa = 3.75QNEE326 pKa = 4.34TLTGRR331 pKa = 11.84FEE333 pKa = 4.0YY334 pKa = 10.45HH335 pKa = 6.68KK336 pKa = 10.77NLSRR340 pKa = 11.84YY341 pKa = 8.62LQVSVRR347 pKa = 11.84GSVGFSADD355 pKa = 3.84TPPKK359 pKa = 10.65SSFQGTNVYY368 pKa = 10.7YY369 pKa = 10.55KK370 pKa = 10.55VLKK373 pKa = 9.98TMIAHH378 pKa = 6.14RR379 pKa = 11.84VDD381 pKa = 4.75RR382 pKa = 11.84FGQQPKK388 pKa = 9.74RR389 pKa = 11.84YY390 pKa = 8.84RR391 pKa = 11.84IIGNPSEE398 pKa = 4.43VVSSFNGTAVDD409 pKa = 4.36LGRR412 pKa = 11.84PAITEE417 pKa = 3.95PVEE420 pKa = 5.34SIIQTLQASTT430 pKa = 3.59
Molecular weight: 48.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
704 |
274 |
430 |
352.0 |
40.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.119 ± 0.511 | 0.71 ± 0.459 |
4.972 ± 0.531 | 4.83 ± 1.511 |
5.54 ± 0.183 | 6.25 ± 0.419 |
1.989 ± 0.346 | 5.966 ± 0.146 |
8.807 ± 0.641 | 3.977 ± 0.246 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.131 ± 0.41 | 5.54 ± 1.38 |
5.256 ± 0.536 | 4.83 ± 0.171 |
5.682 ± 0.127 | 7.813 ± 2.101 |
7.528 ± 0.81 | 7.386 ± 0.053 |
1.847 ± 0.657 | 4.83 ± 0.618 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
