
Nariva virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Narmovirus; Nariva narmovirus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
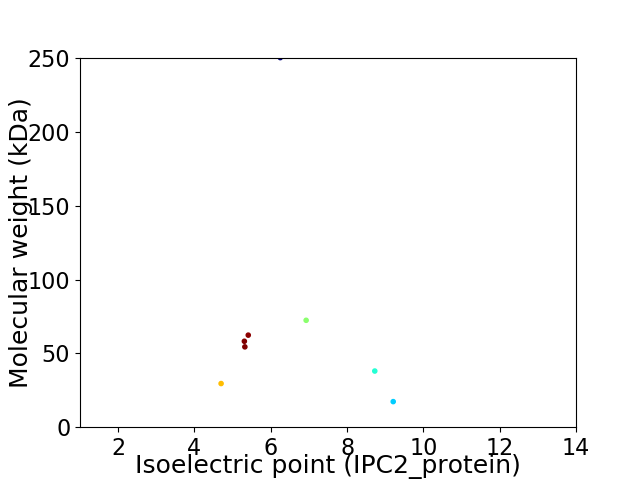
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B8XH61|B8XH61_9MONO Isoform of B8XH59 C protein OS=Nariva virus OX=590647 GN=P/V/C PE=4 SV=1
MM1 pKa = 6.91EE2 pKa = 4.72TNGQKK7 pKa = 10.37VIRR10 pKa = 11.84DD11 pKa = 3.98ALQVLAVVKK20 pKa = 9.18EE21 pKa = 4.31TDD23 pKa = 3.28SPTTEE28 pKa = 4.11NQCGRR33 pKa = 11.84RR34 pKa = 11.84LSVQGVTTQTSTTDD48 pKa = 3.42PEE50 pKa = 4.66TKK52 pKa = 10.41GEE54 pKa = 4.16SGSEE58 pKa = 4.12EE59 pKa = 3.92EE60 pKa = 5.62SGSGWGGEE68 pKa = 4.3GNSSQEE74 pKa = 3.96ASGSNQLVDD83 pKa = 3.04NAEE86 pKa = 4.12RR87 pKa = 11.84YY88 pKa = 10.31GEE90 pKa = 4.46GEE92 pKa = 4.1SKK94 pKa = 10.84GPQGPGGFDD103 pKa = 3.39GVHH106 pKa = 7.13DD107 pKa = 4.02SPVRR111 pKa = 11.84QDD113 pKa = 4.23HH114 pKa = 6.68PDD116 pKa = 3.0EE117 pKa = 4.96GIRR120 pKa = 11.84RR121 pKa = 11.84EE122 pKa = 3.98PCYY125 pKa = 10.92VLLGAASGGAYY136 pKa = 10.21LSEE139 pKa = 4.56SEE141 pKa = 5.11GEE143 pKa = 4.21GSPTRR148 pKa = 11.84GSPVGEE154 pKa = 4.17SGDD157 pKa = 3.66AKK159 pKa = 10.91VIAPPNRR166 pKa = 11.84GSTINDD172 pKa = 3.34NPSCDD177 pKa = 3.77DD178 pKa = 3.51VKK180 pKa = 11.13EE181 pKa = 4.14SLGFISQPKK190 pKa = 8.34PRR192 pKa = 11.84RR193 pKa = 11.84LTGIDD198 pKa = 3.15TSTDD202 pKa = 2.94TGTPIPPPRR211 pKa = 11.84DD212 pKa = 3.33YY213 pKa = 10.88TKK215 pKa = 9.98RR216 pKa = 11.84GHH218 pKa = 5.71RR219 pKa = 11.84RR220 pKa = 11.84EE221 pKa = 4.26YY222 pKa = 10.64SLTWTDD228 pKa = 3.37CGFLVEE234 pKa = 4.63SWCNPVCARR243 pKa = 11.84VTPLPRR249 pKa = 11.84RR250 pKa = 11.84EE251 pKa = 3.88SCKK254 pKa = 10.38CGKK257 pKa = 10.43CPVFCPEE264 pKa = 4.07CVGPGVSQEE273 pKa = 5.06DD274 pKa = 4.41IYY276 pKa = 11.86NDD278 pKa = 3.29
MM1 pKa = 6.91EE2 pKa = 4.72TNGQKK7 pKa = 10.37VIRR10 pKa = 11.84DD11 pKa = 3.98ALQVLAVVKK20 pKa = 9.18EE21 pKa = 4.31TDD23 pKa = 3.28SPTTEE28 pKa = 4.11NQCGRR33 pKa = 11.84RR34 pKa = 11.84LSVQGVTTQTSTTDD48 pKa = 3.42PEE50 pKa = 4.66TKK52 pKa = 10.41GEE54 pKa = 4.16SGSEE58 pKa = 4.12EE59 pKa = 3.92EE60 pKa = 5.62SGSGWGGEE68 pKa = 4.3GNSSQEE74 pKa = 3.96ASGSNQLVDD83 pKa = 3.04NAEE86 pKa = 4.12RR87 pKa = 11.84YY88 pKa = 10.31GEE90 pKa = 4.46GEE92 pKa = 4.1SKK94 pKa = 10.84GPQGPGGFDD103 pKa = 3.39GVHH106 pKa = 7.13DD107 pKa = 4.02SPVRR111 pKa = 11.84QDD113 pKa = 4.23HH114 pKa = 6.68PDD116 pKa = 3.0EE117 pKa = 4.96GIRR120 pKa = 11.84RR121 pKa = 11.84EE122 pKa = 3.98PCYY125 pKa = 10.92VLLGAASGGAYY136 pKa = 10.21LSEE139 pKa = 4.56SEE141 pKa = 5.11GEE143 pKa = 4.21GSPTRR148 pKa = 11.84GSPVGEE154 pKa = 4.17SGDD157 pKa = 3.66AKK159 pKa = 10.91VIAPPNRR166 pKa = 11.84GSTINDD172 pKa = 3.34NPSCDD177 pKa = 3.77DD178 pKa = 3.51VKK180 pKa = 11.13EE181 pKa = 4.14SLGFISQPKK190 pKa = 8.34PRR192 pKa = 11.84RR193 pKa = 11.84LTGIDD198 pKa = 3.15TSTDD202 pKa = 2.94TGTPIPPPRR211 pKa = 11.84DD212 pKa = 3.33YY213 pKa = 10.88TKK215 pKa = 9.98RR216 pKa = 11.84GHH218 pKa = 5.71RR219 pKa = 11.84RR220 pKa = 11.84EE221 pKa = 4.26YY222 pKa = 10.64SLTWTDD228 pKa = 3.37CGFLVEE234 pKa = 4.63SWCNPVCARR243 pKa = 11.84VTPLPRR249 pKa = 11.84RR250 pKa = 11.84EE251 pKa = 3.88SCKK254 pKa = 10.38CGKK257 pKa = 10.43CPVFCPEE264 pKa = 4.07CVGPGVSQEE273 pKa = 5.06DD274 pKa = 4.41IYY276 pKa = 11.86NDD278 pKa = 3.29
Molecular weight: 29.57 kDa
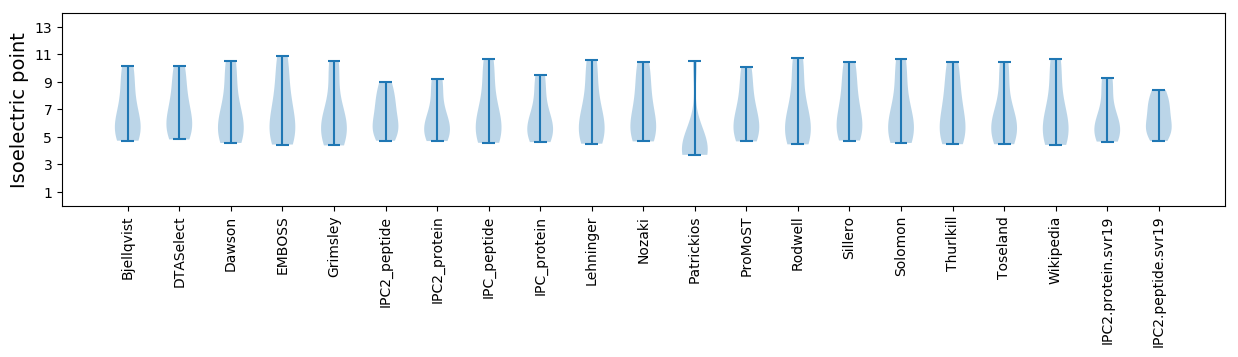
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B8XH61|B8XH61_9MONO Isoform of B8XH59 C protein OS=Nariva virus OX=590647 GN=P/V/C PE=4 SV=1
MM1 pKa = 8.09PSRR4 pKa = 11.84FLQSLRR10 pKa = 11.84KK11 pKa = 8.48LTLRR15 pKa = 11.84RR16 pKa = 11.84QRR18 pKa = 11.84TNAEE22 pKa = 3.72EE23 pKa = 4.52DD24 pKa = 3.65SQSRR28 pKa = 11.84EE29 pKa = 4.17SPPRR33 pKa = 11.84PPPRR37 pKa = 11.84TPRR40 pKa = 11.84PRR42 pKa = 11.84VRR44 pKa = 11.84VGVRR48 pKa = 11.84KK49 pKa = 10.11NPDD52 pKa = 2.93LVGVEE57 pKa = 4.93KK58 pKa = 9.73EE59 pKa = 4.19TQARR63 pKa = 11.84KK64 pKa = 8.44QAEE67 pKa = 4.73AINLLTMLRR76 pKa = 11.84DD77 pKa = 3.6MEE79 pKa = 4.37RR80 pKa = 11.84EE81 pKa = 3.71NLRR84 pKa = 11.84GLRR87 pKa = 11.84GLEE90 pKa = 3.76GLTEE94 pKa = 4.05YY95 pKa = 8.12TTVQFVKK102 pKa = 9.61TILMRR107 pKa = 11.84VSEE110 pKa = 4.45GSPVTSCWVQQVEE123 pKa = 4.1EE124 pKa = 5.36HH125 pKa = 6.33ICQSQRR131 pKa = 11.84EE132 pKa = 4.23KK133 pKa = 10.94EE134 pKa = 4.19ALHH137 pKa = 6.7EE138 pKa = 4.1AVQWVRR144 pKa = 11.84AVMQKK149 pKa = 10.74
MM1 pKa = 8.09PSRR4 pKa = 11.84FLQSLRR10 pKa = 11.84KK11 pKa = 8.48LTLRR15 pKa = 11.84RR16 pKa = 11.84QRR18 pKa = 11.84TNAEE22 pKa = 3.72EE23 pKa = 4.52DD24 pKa = 3.65SQSRR28 pKa = 11.84EE29 pKa = 4.17SPPRR33 pKa = 11.84PPPRR37 pKa = 11.84TPRR40 pKa = 11.84PRR42 pKa = 11.84VRR44 pKa = 11.84VGVRR48 pKa = 11.84KK49 pKa = 10.11NPDD52 pKa = 2.93LVGVEE57 pKa = 4.93KK58 pKa = 9.73EE59 pKa = 4.19TQARR63 pKa = 11.84KK64 pKa = 8.44QAEE67 pKa = 4.73AINLLTMLRR76 pKa = 11.84DD77 pKa = 3.6MEE79 pKa = 4.37RR80 pKa = 11.84EE81 pKa = 3.71NLRR84 pKa = 11.84GLRR87 pKa = 11.84GLEE90 pKa = 3.76GLTEE94 pKa = 4.05YY95 pKa = 8.12TTVQFVKK102 pKa = 9.61TILMRR107 pKa = 11.84VSEE110 pKa = 4.45GSPVTSCWVQQVEE123 pKa = 4.1EE124 pKa = 5.36HH125 pKa = 6.33ICQSQRR131 pKa = 11.84EE132 pKa = 4.23KK133 pKa = 10.94EE134 pKa = 4.19ALHH137 pKa = 6.7EE138 pKa = 4.1AVQWVRR144 pKa = 11.84AVMQKK149 pKa = 10.74
Molecular weight: 17.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5226 |
149 |
2201 |
653.3 |
72.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.487 ± 0.542 | 1.933 ± 0.25 |
5.549 ± 0.317 | 5.434 ± 0.481 |
2.832 ± 0.401 | 6.525 ± 0.779 |
2.354 ± 0.479 | 7.08 ± 0.795 |
4.363 ± 0.26 | 9.127 ± 0.656 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.698 ± 0.354 | 4.497 ± 0.259 |
5.683 ± 0.612 | 4.286 ± 0.484 |
6.008 ± 0.317 | 7.597 ± 0.515 |
6.142 ± 0.364 | 6.774 ± 0.368 |
1.11 ± 0.169 | 3.521 ± 0.402 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
