
Hubei narna-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
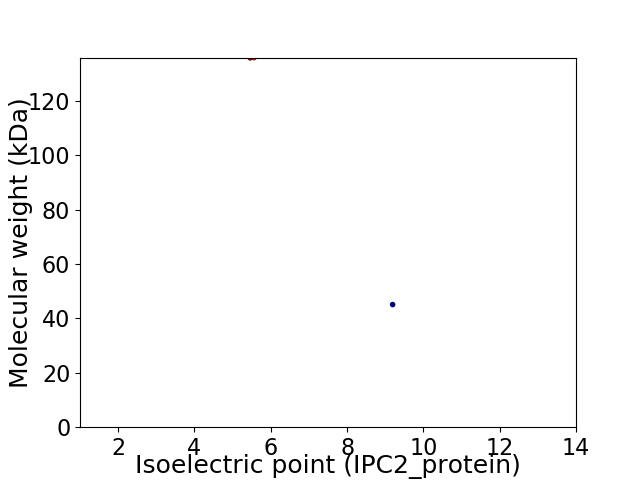
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIN3|A0A1L3KIN3_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 11 OX=1922941 PE=4 SV=1
MM1 pKa = 7.79ISPFHH6 pKa = 6.66ASDD9 pKa = 3.42VKK11 pKa = 11.21SNSFTSLLTYY21 pKa = 10.79VDD23 pKa = 4.94LLCDD27 pKa = 4.62FNPKK31 pKa = 9.68GRR33 pKa = 11.84LRR35 pKa = 11.84VPLVPLLGRR44 pKa = 11.84ISLSDD49 pKa = 4.2LYY51 pKa = 10.89TKK53 pKa = 10.08EE54 pKa = 3.93EE55 pKa = 4.33WNQSDD60 pKa = 3.82TFYY63 pKa = 11.21LIGPHH68 pKa = 6.45GSDD71 pKa = 5.01SITTNPTLLSALNLRR86 pKa = 11.84TLFPADD92 pKa = 3.99CGPVEE97 pKa = 4.05HH98 pKa = 7.09SLFFGPHH105 pKa = 5.48SPFPPGTTFQYY116 pKa = 10.61IPGGSSYY123 pKa = 12.06SMLFFLRR130 pKa = 11.84SILEE134 pKa = 3.96VYY136 pKa = 10.42IDD138 pKa = 4.89FFTSDD143 pKa = 3.82LVTPSMRR150 pKa = 11.84DD151 pKa = 2.7RR152 pKa = 11.84FLAYY156 pKa = 10.19QPTSQSACIAYY167 pKa = 10.11LSSLGSSFVKK177 pKa = 10.22VVKK180 pKa = 10.38FYY182 pKa = 10.13TAYY185 pKa = 10.12PMAAFLRR192 pKa = 11.84QEE194 pKa = 4.39DD195 pKa = 4.18MPPRR199 pKa = 11.84PSQIPEE205 pKa = 3.55VRR207 pKa = 11.84TWLWDD212 pKa = 3.1GDD214 pKa = 3.54FRR216 pKa = 11.84KK217 pKa = 9.54FLKK220 pKa = 10.67QITLYY225 pKa = 10.61RR226 pKa = 11.84NPEE229 pKa = 4.03YY230 pKa = 10.95APLIWGTLQGVKK242 pKa = 9.96RR243 pKa = 11.84GCASVPDD250 pKa = 4.11TFVSDD255 pKa = 3.98SYY257 pKa = 11.46ASHH260 pKa = 6.8FRR262 pKa = 11.84TMAMEE267 pKa = 4.26SRR269 pKa = 11.84NSLLQEE275 pKa = 3.96KK276 pKa = 10.39ALDD279 pKa = 3.75LKK281 pKa = 10.96EE282 pKa = 4.47ILLNPDD288 pKa = 2.69NVLPLWGDD296 pKa = 3.43EE297 pKa = 4.07NEE299 pKa = 4.35RR300 pKa = 11.84ASEE303 pKa = 4.1LEE305 pKa = 3.93EE306 pKa = 4.24EE307 pKa = 4.49IKK309 pKa = 10.68ILEE312 pKa = 4.69DD313 pKa = 2.9KK314 pKa = 10.77TGIRR318 pKa = 11.84NTRR321 pKa = 11.84AKK323 pKa = 9.53ATLVFNILPPGSDD336 pKa = 3.75FNGTIYY342 pKa = 10.63RR343 pKa = 11.84PIKK346 pKa = 9.97MEE348 pKa = 4.16EE349 pKa = 3.71FDD351 pKa = 5.32ALDD354 pKa = 3.78SPIEE358 pKa = 4.17PSSSASYY365 pKa = 9.98RR366 pKa = 11.84YY367 pKa = 9.74SSRR370 pKa = 11.84KK371 pKa = 8.71GGQRR375 pKa = 11.84MDD377 pKa = 3.19VMQEE381 pKa = 3.53VMPPVSLPVSFPTPPSGDD399 pKa = 3.62LEE401 pKa = 5.1LNLLDD406 pKa = 5.59AIDD409 pKa = 3.9VGLVNSFSEE418 pKa = 4.3LEE420 pKa = 4.02SYY422 pKa = 10.56YY423 pKa = 10.35RR424 pKa = 11.84QQLSMGDD431 pKa = 3.37VLRR434 pKa = 11.84SVRR437 pKa = 11.84QSHH440 pKa = 7.54PDD442 pKa = 3.01HH443 pKa = 6.78QYY445 pKa = 10.2IRR447 pKa = 11.84DD448 pKa = 3.57WYY450 pKa = 10.67SGVTRR455 pKa = 11.84SWSTDD460 pKa = 3.02DD461 pKa = 4.48LRR463 pKa = 11.84YY464 pKa = 10.08SLSSILRR471 pKa = 11.84VFCFRR476 pKa = 11.84LPLTYY481 pKa = 10.64SVDD484 pKa = 3.73DD485 pKa = 4.01FLFASEE491 pKa = 4.63PPFVIPRR498 pKa = 11.84LLSQLPDD505 pKa = 5.22GIPQLISSFLGYY517 pKa = 8.76TVTPEE522 pKa = 4.11EE523 pKa = 4.13EE524 pKa = 4.16SHH526 pKa = 5.05VVKK529 pKa = 10.95VSSAYY534 pKa = 9.64TEE536 pKa = 3.57MKK538 pKa = 9.3MRR540 pKa = 11.84SEE542 pKa = 4.09NQEE545 pKa = 3.29ISSILSSITNGVTYY559 pKa = 10.29DD560 pKa = 3.88PSFGDD565 pKa = 3.73IDD567 pKa = 3.84VGLQKK572 pKa = 10.34MFVTDD577 pKa = 4.07PNSVHH582 pKa = 7.22EE583 pKa = 4.56IRR585 pKa = 11.84GHH587 pKa = 5.94LPAFFPYY594 pKa = 9.7TSPRR598 pKa = 11.84RR599 pKa = 11.84YY600 pKa = 9.99LLSLQQLQPDD610 pKa = 3.9IPVEE614 pKa = 4.05ANVIALIEE622 pKa = 4.0PLKK625 pKa = 10.84VRR627 pKa = 11.84VITTGEE633 pKa = 3.44AAAYY637 pKa = 8.97YY638 pKa = 10.04LSKK641 pKa = 10.48PLQRR645 pKa = 11.84SFWKK649 pKa = 10.25HH650 pKa = 6.07LYY652 pKa = 9.75QFPQFVLTGQPLTVEE667 pKa = 4.3ILDD670 pKa = 3.72QLEE673 pKa = 4.46DD674 pKa = 3.51STLAFCAKK682 pKa = 10.28YY683 pKa = 10.36SVPTLDD689 pKa = 2.95QWVSGDD695 pKa = 3.68YY696 pKa = 10.65KK697 pKa = 11.11GATDD701 pKa = 3.73TLDD704 pKa = 3.6ILSSMASFSAGINVLSQHH722 pKa = 5.76SQLSEE727 pKa = 3.95QFEE730 pKa = 4.36KK731 pKa = 11.18LLDD734 pKa = 3.98LNRR737 pKa = 11.84QVLDD741 pKa = 3.46RR742 pKa = 11.84HH743 pKa = 5.3TLNYY747 pKa = 9.8PKK749 pKa = 10.04GTLSSLTHH757 pKa = 5.85DD758 pKa = 4.03QIVEE762 pKa = 3.78LGGYY766 pKa = 8.9YY767 pKa = 10.7GEE769 pKa = 4.33NGEE772 pKa = 4.37VKK774 pKa = 10.46LLQGTGQLMGSPLSFPILCLINFIAYY800 pKa = 5.19WTSMEE805 pKa = 4.4KK806 pKa = 10.71YY807 pKa = 9.71LGRR810 pKa = 11.84EE811 pKa = 3.73IPDD814 pKa = 3.23FWEE817 pKa = 4.23LPVRR821 pKa = 11.84INGDD825 pKa = 3.78DD826 pKa = 3.57ILFKK830 pKa = 11.25SNTEE834 pKa = 4.36HH835 pKa = 6.19YY836 pKa = 10.11SIWKK840 pKa = 10.01EE841 pKa = 3.78EE842 pKa = 4.23VKK844 pKa = 10.69VHH846 pKa = 6.08SFEE849 pKa = 5.38LSMGKK854 pKa = 9.91NYY856 pKa = 9.88IHH858 pKa = 7.13PRR860 pKa = 11.84FLTVNSVLYY869 pKa = 10.51YY870 pKa = 7.41YY871 pKa = 9.82TRR873 pKa = 11.84HH874 pKa = 6.47HH875 pKa = 7.38DD876 pKa = 3.59GSPADD881 pKa = 4.08FGKK884 pKa = 10.67VPYY887 pKa = 10.73LNVGLLLSQSKK898 pKa = 10.37GVDD901 pKa = 3.06RR902 pKa = 11.84DD903 pKa = 3.54PQRR906 pKa = 11.84RR907 pKa = 11.84LPFAEE912 pKa = 5.15LYY914 pKa = 9.95RR915 pKa = 11.84ISVGDD920 pKa = 3.37SCNVRR925 pKa = 11.84RR926 pKa = 11.84SHH928 pKa = 7.48DD929 pKa = 3.35RR930 pKa = 11.84FIHH933 pKa = 5.55YY934 pKa = 9.73NIKK937 pKa = 10.18QIKK940 pKa = 9.15QFTNNGQFNLFIPVHH955 pKa = 5.93LGGLGFPLLPLIRR968 pKa = 11.84DD969 pKa = 3.29WRR971 pKa = 11.84VPSTDD976 pKa = 2.66GGTVFKK982 pKa = 11.35VSFTKK987 pKa = 10.37FQIQFATFLSMRR999 pKa = 11.84VEE1001 pKa = 4.33QVSLTGEE1008 pKa = 3.61IPYY1011 pKa = 10.15KK1012 pKa = 10.68YY1013 pKa = 9.79FAALVPQDD1021 pKa = 3.72LASYY1025 pKa = 9.65MRR1027 pKa = 11.84DD1028 pKa = 3.04LNTARR1033 pKa = 11.84KK1034 pKa = 9.59KK1035 pKa = 9.77IGKK1038 pKa = 9.28SEE1040 pKa = 3.98VLPPAGALSPVMYY1053 pKa = 7.95YY1054 pKa = 7.45TWKK1057 pKa = 10.48GLRR1060 pKa = 11.84PLQPLYY1066 pKa = 10.75LPSGFDD1072 pKa = 3.7LSSSTIFKK1080 pKa = 10.41DD1081 pKa = 2.99IMYY1084 pKa = 9.79VEE1086 pKa = 4.6PSSIKK1091 pKa = 9.61RR1092 pKa = 11.84TLLSYY1097 pKa = 10.78SYY1099 pKa = 9.54DD1100 pKa = 3.33TSLPLQYY1107 pKa = 10.34RR1108 pKa = 11.84LPSRR1112 pKa = 11.84SVMSEE1117 pKa = 3.77FSSFFMKK1124 pKa = 10.49GDD1126 pKa = 3.31EE1127 pKa = 4.39KK1128 pKa = 11.36GRR1130 pKa = 11.84FSLPLISSEE1139 pKa = 3.76RR1140 pKa = 11.84LLSNEE1145 pKa = 3.49QRR1147 pKa = 11.84PVYY1150 pKa = 10.2VDD1152 pKa = 4.55RR1153 pKa = 11.84EE1154 pKa = 4.21ALHH1157 pKa = 6.13QLFSTTGLSDD1167 pKa = 3.61LMGDD1171 pKa = 3.86HH1172 pKa = 6.66GPSVFLEE1179 pKa = 4.12LADD1182 pKa = 5.15LNNLPHH1188 pKa = 6.36TPIHH1192 pKa = 6.82LDD1194 pKa = 3.33YY1195 pKa = 10.99FLL1197 pKa = 6.1
MM1 pKa = 7.79ISPFHH6 pKa = 6.66ASDD9 pKa = 3.42VKK11 pKa = 11.21SNSFTSLLTYY21 pKa = 10.79VDD23 pKa = 4.94LLCDD27 pKa = 4.62FNPKK31 pKa = 9.68GRR33 pKa = 11.84LRR35 pKa = 11.84VPLVPLLGRR44 pKa = 11.84ISLSDD49 pKa = 4.2LYY51 pKa = 10.89TKK53 pKa = 10.08EE54 pKa = 3.93EE55 pKa = 4.33WNQSDD60 pKa = 3.82TFYY63 pKa = 11.21LIGPHH68 pKa = 6.45GSDD71 pKa = 5.01SITTNPTLLSALNLRR86 pKa = 11.84TLFPADD92 pKa = 3.99CGPVEE97 pKa = 4.05HH98 pKa = 7.09SLFFGPHH105 pKa = 5.48SPFPPGTTFQYY116 pKa = 10.61IPGGSSYY123 pKa = 12.06SMLFFLRR130 pKa = 11.84SILEE134 pKa = 3.96VYY136 pKa = 10.42IDD138 pKa = 4.89FFTSDD143 pKa = 3.82LVTPSMRR150 pKa = 11.84DD151 pKa = 2.7RR152 pKa = 11.84FLAYY156 pKa = 10.19QPTSQSACIAYY167 pKa = 10.11LSSLGSSFVKK177 pKa = 10.22VVKK180 pKa = 10.38FYY182 pKa = 10.13TAYY185 pKa = 10.12PMAAFLRR192 pKa = 11.84QEE194 pKa = 4.39DD195 pKa = 4.18MPPRR199 pKa = 11.84PSQIPEE205 pKa = 3.55VRR207 pKa = 11.84TWLWDD212 pKa = 3.1GDD214 pKa = 3.54FRR216 pKa = 11.84KK217 pKa = 9.54FLKK220 pKa = 10.67QITLYY225 pKa = 10.61RR226 pKa = 11.84NPEE229 pKa = 4.03YY230 pKa = 10.95APLIWGTLQGVKK242 pKa = 9.96RR243 pKa = 11.84GCASVPDD250 pKa = 4.11TFVSDD255 pKa = 3.98SYY257 pKa = 11.46ASHH260 pKa = 6.8FRR262 pKa = 11.84TMAMEE267 pKa = 4.26SRR269 pKa = 11.84NSLLQEE275 pKa = 3.96KK276 pKa = 10.39ALDD279 pKa = 3.75LKK281 pKa = 10.96EE282 pKa = 4.47ILLNPDD288 pKa = 2.69NVLPLWGDD296 pKa = 3.43EE297 pKa = 4.07NEE299 pKa = 4.35RR300 pKa = 11.84ASEE303 pKa = 4.1LEE305 pKa = 3.93EE306 pKa = 4.24EE307 pKa = 4.49IKK309 pKa = 10.68ILEE312 pKa = 4.69DD313 pKa = 2.9KK314 pKa = 10.77TGIRR318 pKa = 11.84NTRR321 pKa = 11.84AKK323 pKa = 9.53ATLVFNILPPGSDD336 pKa = 3.75FNGTIYY342 pKa = 10.63RR343 pKa = 11.84PIKK346 pKa = 9.97MEE348 pKa = 4.16EE349 pKa = 3.71FDD351 pKa = 5.32ALDD354 pKa = 3.78SPIEE358 pKa = 4.17PSSSASYY365 pKa = 9.98RR366 pKa = 11.84YY367 pKa = 9.74SSRR370 pKa = 11.84KK371 pKa = 8.71GGQRR375 pKa = 11.84MDD377 pKa = 3.19VMQEE381 pKa = 3.53VMPPVSLPVSFPTPPSGDD399 pKa = 3.62LEE401 pKa = 5.1LNLLDD406 pKa = 5.59AIDD409 pKa = 3.9VGLVNSFSEE418 pKa = 4.3LEE420 pKa = 4.02SYY422 pKa = 10.56YY423 pKa = 10.35RR424 pKa = 11.84QQLSMGDD431 pKa = 3.37VLRR434 pKa = 11.84SVRR437 pKa = 11.84QSHH440 pKa = 7.54PDD442 pKa = 3.01HH443 pKa = 6.78QYY445 pKa = 10.2IRR447 pKa = 11.84DD448 pKa = 3.57WYY450 pKa = 10.67SGVTRR455 pKa = 11.84SWSTDD460 pKa = 3.02DD461 pKa = 4.48LRR463 pKa = 11.84YY464 pKa = 10.08SLSSILRR471 pKa = 11.84VFCFRR476 pKa = 11.84LPLTYY481 pKa = 10.64SVDD484 pKa = 3.73DD485 pKa = 4.01FLFASEE491 pKa = 4.63PPFVIPRR498 pKa = 11.84LLSQLPDD505 pKa = 5.22GIPQLISSFLGYY517 pKa = 8.76TVTPEE522 pKa = 4.11EE523 pKa = 4.13EE524 pKa = 4.16SHH526 pKa = 5.05VVKK529 pKa = 10.95VSSAYY534 pKa = 9.64TEE536 pKa = 3.57MKK538 pKa = 9.3MRR540 pKa = 11.84SEE542 pKa = 4.09NQEE545 pKa = 3.29ISSILSSITNGVTYY559 pKa = 10.29DD560 pKa = 3.88PSFGDD565 pKa = 3.73IDD567 pKa = 3.84VGLQKK572 pKa = 10.34MFVTDD577 pKa = 4.07PNSVHH582 pKa = 7.22EE583 pKa = 4.56IRR585 pKa = 11.84GHH587 pKa = 5.94LPAFFPYY594 pKa = 9.7TSPRR598 pKa = 11.84RR599 pKa = 11.84YY600 pKa = 9.99LLSLQQLQPDD610 pKa = 3.9IPVEE614 pKa = 4.05ANVIALIEE622 pKa = 4.0PLKK625 pKa = 10.84VRR627 pKa = 11.84VITTGEE633 pKa = 3.44AAAYY637 pKa = 8.97YY638 pKa = 10.04LSKK641 pKa = 10.48PLQRR645 pKa = 11.84SFWKK649 pKa = 10.25HH650 pKa = 6.07LYY652 pKa = 9.75QFPQFVLTGQPLTVEE667 pKa = 4.3ILDD670 pKa = 3.72QLEE673 pKa = 4.46DD674 pKa = 3.51STLAFCAKK682 pKa = 10.28YY683 pKa = 10.36SVPTLDD689 pKa = 2.95QWVSGDD695 pKa = 3.68YY696 pKa = 10.65KK697 pKa = 11.11GATDD701 pKa = 3.73TLDD704 pKa = 3.6ILSSMASFSAGINVLSQHH722 pKa = 5.76SQLSEE727 pKa = 3.95QFEE730 pKa = 4.36KK731 pKa = 11.18LLDD734 pKa = 3.98LNRR737 pKa = 11.84QVLDD741 pKa = 3.46RR742 pKa = 11.84HH743 pKa = 5.3TLNYY747 pKa = 9.8PKK749 pKa = 10.04GTLSSLTHH757 pKa = 5.85DD758 pKa = 4.03QIVEE762 pKa = 3.78LGGYY766 pKa = 8.9YY767 pKa = 10.7GEE769 pKa = 4.33NGEE772 pKa = 4.37VKK774 pKa = 10.46LLQGTGQLMGSPLSFPILCLINFIAYY800 pKa = 5.19WTSMEE805 pKa = 4.4KK806 pKa = 10.71YY807 pKa = 9.71LGRR810 pKa = 11.84EE811 pKa = 3.73IPDD814 pKa = 3.23FWEE817 pKa = 4.23LPVRR821 pKa = 11.84INGDD825 pKa = 3.78DD826 pKa = 3.57ILFKK830 pKa = 11.25SNTEE834 pKa = 4.36HH835 pKa = 6.19YY836 pKa = 10.11SIWKK840 pKa = 10.01EE841 pKa = 3.78EE842 pKa = 4.23VKK844 pKa = 10.69VHH846 pKa = 6.08SFEE849 pKa = 5.38LSMGKK854 pKa = 9.91NYY856 pKa = 9.88IHH858 pKa = 7.13PRR860 pKa = 11.84FLTVNSVLYY869 pKa = 10.51YY870 pKa = 7.41YY871 pKa = 9.82TRR873 pKa = 11.84HH874 pKa = 6.47HH875 pKa = 7.38DD876 pKa = 3.59GSPADD881 pKa = 4.08FGKK884 pKa = 10.67VPYY887 pKa = 10.73LNVGLLLSQSKK898 pKa = 10.37GVDD901 pKa = 3.06RR902 pKa = 11.84DD903 pKa = 3.54PQRR906 pKa = 11.84RR907 pKa = 11.84LPFAEE912 pKa = 5.15LYY914 pKa = 9.95RR915 pKa = 11.84ISVGDD920 pKa = 3.37SCNVRR925 pKa = 11.84RR926 pKa = 11.84SHH928 pKa = 7.48DD929 pKa = 3.35RR930 pKa = 11.84FIHH933 pKa = 5.55YY934 pKa = 9.73NIKK937 pKa = 10.18QIKK940 pKa = 9.15QFTNNGQFNLFIPVHH955 pKa = 5.93LGGLGFPLLPLIRR968 pKa = 11.84DD969 pKa = 3.29WRR971 pKa = 11.84VPSTDD976 pKa = 2.66GGTVFKK982 pKa = 11.35VSFTKK987 pKa = 10.37FQIQFATFLSMRR999 pKa = 11.84VEE1001 pKa = 4.33QVSLTGEE1008 pKa = 3.61IPYY1011 pKa = 10.15KK1012 pKa = 10.68YY1013 pKa = 9.79FAALVPQDD1021 pKa = 3.72LASYY1025 pKa = 9.65MRR1027 pKa = 11.84DD1028 pKa = 3.04LNTARR1033 pKa = 11.84KK1034 pKa = 9.59KK1035 pKa = 9.77IGKK1038 pKa = 9.28SEE1040 pKa = 3.98VLPPAGALSPVMYY1053 pKa = 7.95YY1054 pKa = 7.45TWKK1057 pKa = 10.48GLRR1060 pKa = 11.84PLQPLYY1066 pKa = 10.75LPSGFDD1072 pKa = 3.7LSSSTIFKK1080 pKa = 10.41DD1081 pKa = 2.99IMYY1084 pKa = 9.79VEE1086 pKa = 4.6PSSIKK1091 pKa = 9.61RR1092 pKa = 11.84TLLSYY1097 pKa = 10.78SYY1099 pKa = 9.54DD1100 pKa = 3.33TSLPLQYY1107 pKa = 10.34RR1108 pKa = 11.84LPSRR1112 pKa = 11.84SVMSEE1117 pKa = 3.77FSSFFMKK1124 pKa = 10.49GDD1126 pKa = 3.31EE1127 pKa = 4.39KK1128 pKa = 11.36GRR1130 pKa = 11.84FSLPLISSEE1139 pKa = 3.76RR1140 pKa = 11.84LLSNEE1145 pKa = 3.49QRR1147 pKa = 11.84PVYY1150 pKa = 10.2VDD1152 pKa = 4.55RR1153 pKa = 11.84EE1154 pKa = 4.21ALHH1157 pKa = 6.13QLFSTTGLSDD1167 pKa = 3.61LMGDD1171 pKa = 3.86HH1172 pKa = 6.66GPSVFLEE1179 pKa = 4.12LADD1182 pKa = 5.15LNNLPHH1188 pKa = 6.36TPIHH1192 pKa = 6.82LDD1194 pKa = 3.33YY1195 pKa = 10.99FLL1197 pKa = 6.1
Molecular weight: 135.81 kDa
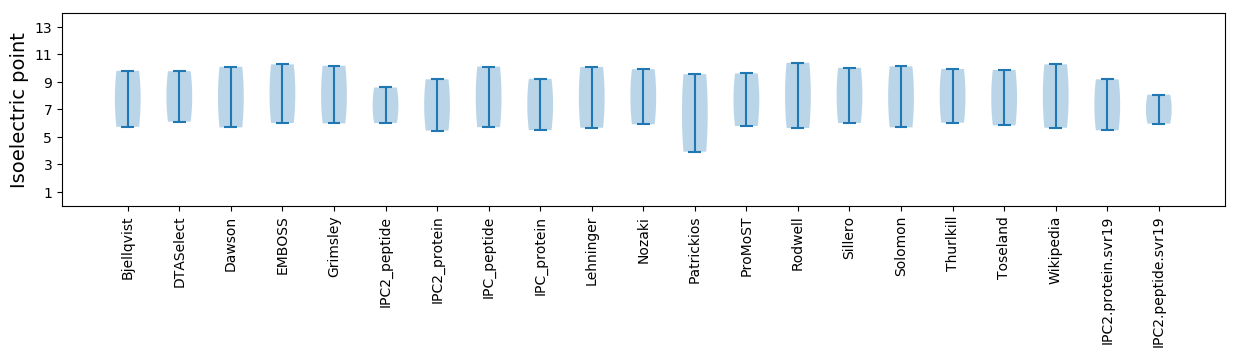
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIN3|A0A1L3KIN3_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 11 OX=1922941 PE=4 SV=1
MM1 pKa = 6.77TRR3 pKa = 11.84SRR5 pKa = 11.84ANNSRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 7.44TRR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84KK18 pKa = 8.45KK19 pKa = 9.86APRR22 pKa = 11.84RR23 pKa = 11.84KK24 pKa = 8.77PRR26 pKa = 11.84SRR28 pKa = 11.84PRR30 pKa = 11.84TSGLSKK36 pKa = 10.91VITDD40 pKa = 3.68YY41 pKa = 11.87AKK43 pKa = 9.92MVMDD47 pKa = 5.16PCHH50 pKa = 5.73STLVRR55 pKa = 11.84TIGTATAGSVTEE67 pKa = 4.5RR68 pKa = 11.84LRR70 pKa = 11.84STIGTPTSAIVSAVGVRR87 pKa = 11.84TIVPAPSGYY96 pKa = 10.12IIWFPSYY103 pKa = 11.28HH104 pKa = 6.45NGTGNLNPANCFIYY118 pKa = 10.07TSPSSSTTTFVVNNDD133 pKa = 3.02GRR135 pKa = 11.84PTNTNALPMGTEE147 pKa = 4.37AYY149 pKa = 7.61DD150 pKa = 3.55TTGLFVQDD158 pKa = 3.96PANNLLSGAASVFSRR173 pKa = 11.84GTTVSACLQLEE184 pKa = 4.27NLVSLSTMQGQVAVVKK200 pKa = 10.31NYY202 pKa = 10.63NLAAFDD208 pKa = 4.83LNSGTNLEE216 pKa = 4.47LKK218 pKa = 10.42APTVNEE224 pKa = 3.69VFAYY228 pKa = 10.15AEE230 pKa = 3.99EE231 pKa = 4.12RR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84IDD236 pKa = 3.34PAGHH240 pKa = 4.96EE241 pKa = 4.73VVWRR245 pKa = 11.84PTDD248 pKa = 3.32ASSVARR254 pKa = 11.84GIGNEE259 pKa = 3.72ALGKK263 pKa = 10.0NFAGSPYY270 pKa = 10.35TDD272 pKa = 2.7VCFQAGQTGINSTNLAATAPEE293 pKa = 4.22HH294 pKa = 6.51QYY296 pKa = 10.95GICIAWRR303 pKa = 11.84GMAAFTTNVGLNNEE317 pKa = 4.95PITLNLIKK325 pKa = 10.4IVDD328 pKa = 4.25FEE330 pKa = 4.2LAARR334 pKa = 11.84NGQIEE339 pKa = 5.15SKK341 pKa = 9.84PKK343 pKa = 9.61PPPAPGVSKK352 pKa = 10.87DD353 pKa = 3.55VGVSFLDD360 pKa = 3.1RR361 pKa = 11.84VFPHH365 pKa = 5.97WQSTTVKK372 pKa = 9.21LAHH375 pKa = 6.79AGAVTAASMIGGSAAGNIVDD395 pKa = 5.82GIITMAQVGDD405 pKa = 3.56VTKK408 pKa = 11.27NKK410 pKa = 10.08IGGKK414 pKa = 9.3NRR416 pKa = 11.84GSQMIADD423 pKa = 4.1YY424 pKa = 11.29
MM1 pKa = 6.77TRR3 pKa = 11.84SRR5 pKa = 11.84ANNSRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 7.44TRR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84KK18 pKa = 8.45KK19 pKa = 9.86APRR22 pKa = 11.84RR23 pKa = 11.84KK24 pKa = 8.77PRR26 pKa = 11.84SRR28 pKa = 11.84PRR30 pKa = 11.84TSGLSKK36 pKa = 10.91VITDD40 pKa = 3.68YY41 pKa = 11.87AKK43 pKa = 9.92MVMDD47 pKa = 5.16PCHH50 pKa = 5.73STLVRR55 pKa = 11.84TIGTATAGSVTEE67 pKa = 4.5RR68 pKa = 11.84LRR70 pKa = 11.84STIGTPTSAIVSAVGVRR87 pKa = 11.84TIVPAPSGYY96 pKa = 10.12IIWFPSYY103 pKa = 11.28HH104 pKa = 6.45NGTGNLNPANCFIYY118 pKa = 10.07TSPSSSTTTFVVNNDD133 pKa = 3.02GRR135 pKa = 11.84PTNTNALPMGTEE147 pKa = 4.37AYY149 pKa = 7.61DD150 pKa = 3.55TTGLFVQDD158 pKa = 3.96PANNLLSGAASVFSRR173 pKa = 11.84GTTVSACLQLEE184 pKa = 4.27NLVSLSTMQGQVAVVKK200 pKa = 10.31NYY202 pKa = 10.63NLAAFDD208 pKa = 4.83LNSGTNLEE216 pKa = 4.47LKK218 pKa = 10.42APTVNEE224 pKa = 3.69VFAYY228 pKa = 10.15AEE230 pKa = 3.99EE231 pKa = 4.12RR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84IDD236 pKa = 3.34PAGHH240 pKa = 4.96EE241 pKa = 4.73VVWRR245 pKa = 11.84PTDD248 pKa = 3.32ASSVARR254 pKa = 11.84GIGNEE259 pKa = 3.72ALGKK263 pKa = 10.0NFAGSPYY270 pKa = 10.35TDD272 pKa = 2.7VCFQAGQTGINSTNLAATAPEE293 pKa = 4.22HH294 pKa = 6.51QYY296 pKa = 10.95GICIAWRR303 pKa = 11.84GMAAFTTNVGLNNEE317 pKa = 4.95PITLNLIKK325 pKa = 10.4IVDD328 pKa = 4.25FEE330 pKa = 4.2LAARR334 pKa = 11.84NGQIEE339 pKa = 5.15SKK341 pKa = 9.84PKK343 pKa = 9.61PPPAPGVSKK352 pKa = 10.87DD353 pKa = 3.55VGVSFLDD360 pKa = 3.1RR361 pKa = 11.84VFPHH365 pKa = 5.97WQSTTVKK372 pKa = 9.21LAHH375 pKa = 6.79AGAVTAASMIGGSAAGNIVDD395 pKa = 5.82GIITMAQVGDD405 pKa = 3.56VTKK408 pKa = 11.27NKK410 pKa = 10.08IGGKK414 pKa = 9.3NRR416 pKa = 11.84GSQMIADD423 pKa = 4.1YY424 pKa = 11.29
Molecular weight: 45.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1621 |
424 |
1197 |
810.5 |
90.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.552 ± 2.544 | 0.802 ± 0.19 |
5.244 ± 0.858 | 4.503 ± 0.723 |
5.12 ± 1.033 | 6.416 ± 1.162 |
1.974 ± 0.281 | 5.12 ± 0.153 |
3.886 ± 0.062 | 10.611 ± 2.489 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.159 ± 0.018 | 4.195 ± 1.448 |
6.909 ± 0.391 | 3.64 ± 0.526 |
5.552 ± 0.41 | 10.056 ± 1.024 |
6.477 ± 1.605 | 6.539 ± 0.744 |
1.11 ± 0.084 | 4.133 ± 0.892 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
