
Rhodospirillales bacterium URHD0017
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; unclassified Rhodospirillales
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
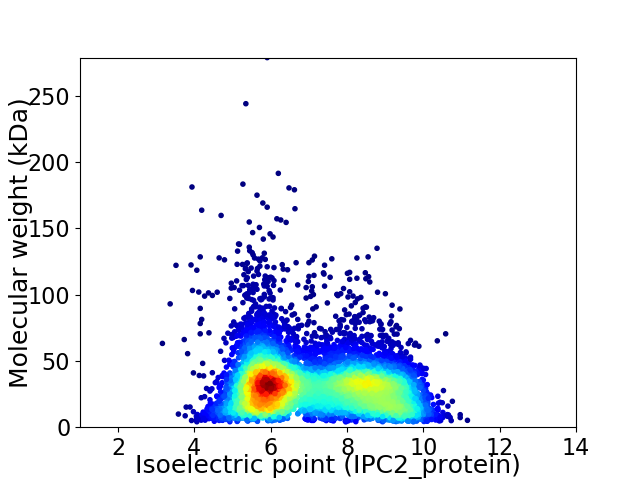
Virtual 2D-PAGE plot for 7953 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H8YBL7|A0A1H8YBL7_9PROT DNA-binding transcriptional regulator LysR family OS=Rhodospirillales bacterium URHD0017 OX=1380357 GN=SAMN02990966_07307 PE=3 SV=1
MM1 pKa = 7.25ATLNVGAGQAYY12 pKa = 6.91ATLSAAVASSHH23 pKa = 6.91DD24 pKa = 3.53GDD26 pKa = 4.43VIAVQAGTYY35 pKa = 10.39VNDD38 pKa = 3.71FATIFNDD45 pKa = 3.25VTIVGVGGMANFVATVAPPNGKK67 pKa = 10.28GILVTQGDD75 pKa = 3.99VTIQNLSFSGVAVADD90 pKa = 3.96ANGAGIRR97 pKa = 11.84YY98 pKa = 9.09EE99 pKa = 4.17GGNLVVIDD107 pKa = 5.19SYY109 pKa = 11.92FHH111 pKa = 7.17DD112 pKa = 3.59NQMNLLANAADD123 pKa = 3.69GTIRR127 pKa = 11.84IVGSEE132 pKa = 4.27FGATRR137 pKa = 11.84SSDD140 pKa = 3.73SLNHH144 pKa = 5.87NLYY147 pKa = 10.18VGSIGSLVIDD157 pKa = 3.31NSYY160 pKa = 10.83FHH162 pKa = 7.5DD163 pKa = 5.13AYY165 pKa = 10.6DD166 pKa = 3.4GHH168 pKa = 6.0QIKK171 pKa = 10.48SRR173 pKa = 11.84AEE175 pKa = 3.81STTITNSRR183 pKa = 11.84IYY185 pKa = 10.5DD186 pKa = 3.53GSGDD190 pKa = 3.49GSYY193 pKa = 10.65TIDD196 pKa = 4.09LPNGGVGVISNNVIEE211 pKa = 4.67QGPHH215 pKa = 5.53SDD217 pKa = 3.32NPFIIAYY224 pKa = 8.82SEE226 pKa = 4.02EE227 pKa = 4.57GATHH231 pKa = 7.25ASNSLVIEE239 pKa = 4.61DD240 pKa = 3.58NVVVNDD246 pKa = 3.65LTGNASLLFNASTITGTIRR265 pKa = 11.84DD266 pKa = 3.69NSVYY270 pKa = 10.81GLTADD275 pKa = 3.62QIVYY279 pKa = 10.55GSATVSGTHH288 pKa = 6.26FLSTHH293 pKa = 5.82PTLDD297 pKa = 3.28TTSGVGGGGTGGGGTGGGGDD317 pKa = 3.48GGGGGSAAIGEE328 pKa = 4.59VTGLSQDD335 pKa = 3.06TGTPGDD341 pKa = 4.91FITSVASQTVHH352 pKa = 4.49GTYY355 pKa = 10.6AGTLANSDD363 pKa = 4.37VIQVSADD370 pKa = 2.7GSTWVGATAGAGTWTADD387 pKa = 3.33VTLLPGEE394 pKa = 4.16HH395 pKa = 6.03TLAVRR400 pKa = 11.84TVDD403 pKa = 3.62DD404 pKa = 4.67AGNIVNGTGHH414 pKa = 7.5AYY416 pKa = 10.27DD417 pKa = 4.84LEE419 pKa = 4.67TGGGGDD425 pKa = 3.66GTVLGTAGADD435 pKa = 3.59TLTGTAGADD444 pKa = 3.32TMTGFAGNDD453 pKa = 3.15EE454 pKa = 4.4YY455 pKa = 11.88VVNNVGDD462 pKa = 3.8VVIEE466 pKa = 3.89ARR468 pKa = 11.84GEE470 pKa = 4.18GTDD473 pKa = 3.49TVLSSVSYY481 pKa = 10.46RR482 pKa = 11.84LAWNQSIEE490 pKa = 4.01NLTLTGDD497 pKa = 3.81GNIDD501 pKa = 3.35GTGNSMNNVLQGSAGDD517 pKa = 3.57NVLRR521 pKa = 11.84GGSGNDD527 pKa = 3.42TLSGGDD533 pKa = 3.68GNDD536 pKa = 3.25VLVGGFGRR544 pKa = 11.84DD545 pKa = 3.18IEE547 pKa = 4.54TGGAGADD554 pKa = 2.91RR555 pKa = 11.84FDD557 pKa = 4.26FNSVNEE563 pKa = 4.31SGTTFATRR571 pKa = 11.84EE572 pKa = 3.85QIMGFEE578 pKa = 4.2QGTDD582 pKa = 3.49AIDD585 pKa = 4.18FSTIDD590 pKa = 3.72ANTAAWGNQAFTFIGDD606 pKa = 3.79DD607 pKa = 3.51AFHH610 pKa = 6.42GVAGEE615 pKa = 4.04LHH617 pKa = 5.68QQTDD621 pKa = 4.03GPNTIVSGDD630 pKa = 3.42INGDD634 pKa = 3.52SVADD638 pKa = 3.89FQIEE642 pKa = 4.22LNGAFTLTPNDD653 pKa = 4.29FILL656 pKa = 5.08
MM1 pKa = 7.25ATLNVGAGQAYY12 pKa = 6.91ATLSAAVASSHH23 pKa = 6.91DD24 pKa = 3.53GDD26 pKa = 4.43VIAVQAGTYY35 pKa = 10.39VNDD38 pKa = 3.71FATIFNDD45 pKa = 3.25VTIVGVGGMANFVATVAPPNGKK67 pKa = 10.28GILVTQGDD75 pKa = 3.99VTIQNLSFSGVAVADD90 pKa = 3.96ANGAGIRR97 pKa = 11.84YY98 pKa = 9.09EE99 pKa = 4.17GGNLVVIDD107 pKa = 5.19SYY109 pKa = 11.92FHH111 pKa = 7.17DD112 pKa = 3.59NQMNLLANAADD123 pKa = 3.69GTIRR127 pKa = 11.84IVGSEE132 pKa = 4.27FGATRR137 pKa = 11.84SSDD140 pKa = 3.73SLNHH144 pKa = 5.87NLYY147 pKa = 10.18VGSIGSLVIDD157 pKa = 3.31NSYY160 pKa = 10.83FHH162 pKa = 7.5DD163 pKa = 5.13AYY165 pKa = 10.6DD166 pKa = 3.4GHH168 pKa = 6.0QIKK171 pKa = 10.48SRR173 pKa = 11.84AEE175 pKa = 3.81STTITNSRR183 pKa = 11.84IYY185 pKa = 10.5DD186 pKa = 3.53GSGDD190 pKa = 3.49GSYY193 pKa = 10.65TIDD196 pKa = 4.09LPNGGVGVISNNVIEE211 pKa = 4.67QGPHH215 pKa = 5.53SDD217 pKa = 3.32NPFIIAYY224 pKa = 8.82SEE226 pKa = 4.02EE227 pKa = 4.57GATHH231 pKa = 7.25ASNSLVIEE239 pKa = 4.61DD240 pKa = 3.58NVVVNDD246 pKa = 3.65LTGNASLLFNASTITGTIRR265 pKa = 11.84DD266 pKa = 3.69NSVYY270 pKa = 10.81GLTADD275 pKa = 3.62QIVYY279 pKa = 10.55GSATVSGTHH288 pKa = 6.26FLSTHH293 pKa = 5.82PTLDD297 pKa = 3.28TTSGVGGGGTGGGGTGGGGDD317 pKa = 3.48GGGGGSAAIGEE328 pKa = 4.59VTGLSQDD335 pKa = 3.06TGTPGDD341 pKa = 4.91FITSVASQTVHH352 pKa = 4.49GTYY355 pKa = 10.6AGTLANSDD363 pKa = 4.37VIQVSADD370 pKa = 2.7GSTWVGATAGAGTWTADD387 pKa = 3.33VTLLPGEE394 pKa = 4.16HH395 pKa = 6.03TLAVRR400 pKa = 11.84TVDD403 pKa = 3.62DD404 pKa = 4.67AGNIVNGTGHH414 pKa = 7.5AYY416 pKa = 10.27DD417 pKa = 4.84LEE419 pKa = 4.67TGGGGDD425 pKa = 3.66GTVLGTAGADD435 pKa = 3.59TLTGTAGADD444 pKa = 3.32TMTGFAGNDD453 pKa = 3.15EE454 pKa = 4.4YY455 pKa = 11.88VVNNVGDD462 pKa = 3.8VVIEE466 pKa = 3.89ARR468 pKa = 11.84GEE470 pKa = 4.18GTDD473 pKa = 3.49TVLSSVSYY481 pKa = 10.46RR482 pKa = 11.84LAWNQSIEE490 pKa = 4.01NLTLTGDD497 pKa = 3.81GNIDD501 pKa = 3.35GTGNSMNNVLQGSAGDD517 pKa = 3.57NVLRR521 pKa = 11.84GGSGNDD527 pKa = 3.42TLSGGDD533 pKa = 3.68GNDD536 pKa = 3.25VLVGGFGRR544 pKa = 11.84DD545 pKa = 3.18IEE547 pKa = 4.54TGGAGADD554 pKa = 2.91RR555 pKa = 11.84FDD557 pKa = 4.26FNSVNEE563 pKa = 4.31SGTTFATRR571 pKa = 11.84EE572 pKa = 3.85QIMGFEE578 pKa = 4.2QGTDD582 pKa = 3.49AIDD585 pKa = 4.18FSTIDD590 pKa = 3.72ANTAAWGNQAFTFIGDD606 pKa = 3.79DD607 pKa = 3.51AFHH610 pKa = 6.42GVAGEE615 pKa = 4.04LHH617 pKa = 5.68QQTDD621 pKa = 4.03GPNTIVSGDD630 pKa = 3.42INGDD634 pKa = 3.52SVADD638 pKa = 3.89FQIEE642 pKa = 4.22LNGAFTLTPNDD653 pKa = 4.29FILL656 pKa = 5.08
Molecular weight: 66.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H8U8S1|A0A1H8U8S1_9PROT Uncharacterized protein OS=Rhodospirillales bacterium URHD0017 OX=1380357 GN=SAMN02990966_03594 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIASRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SGRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.09
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIASRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SGRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.09
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2442273 |
39 |
2555 |
307.1 |
33.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.515 ± 0.038 | 0.901 ± 0.008 |
5.535 ± 0.022 | 5.186 ± 0.024 |
3.685 ± 0.015 | 8.723 ± 0.036 |
2.079 ± 0.014 | 4.764 ± 0.018 |
3.562 ± 0.028 | 10.025 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.489 ± 0.013 | 2.574 ± 0.017 |
5.568 ± 0.021 | 3.137 ± 0.016 |
7.3 ± 0.031 | 5.137 ± 0.02 |
5.263 ± 0.021 | 7.691 ± 0.023 |
1.552 ± 0.011 | 2.314 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
