
Glycine max (Soybean) (Glycine hispida)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; 50 kb inversion clade; NPAAA clade;
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
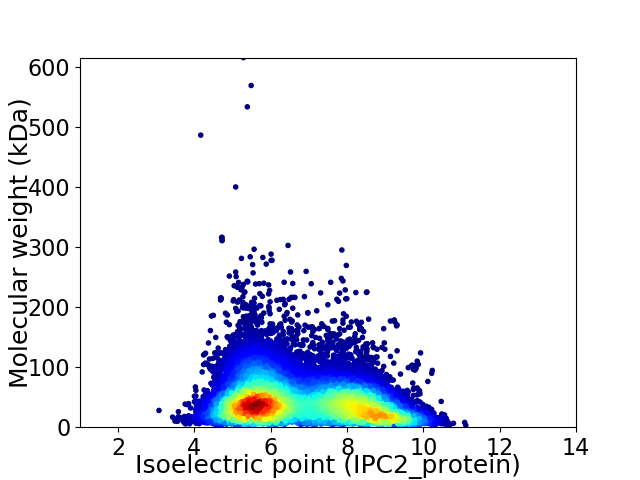
Virtual 2D-PAGE plot for 74868 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
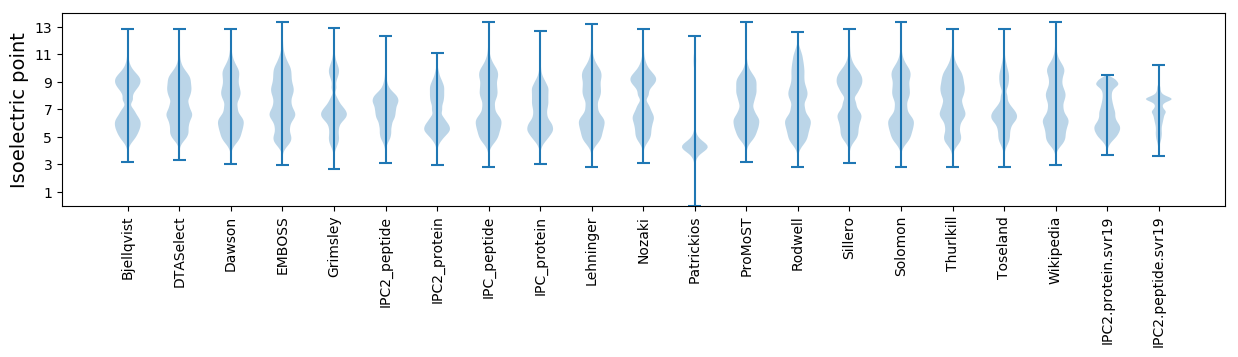
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7MMY5|K7MMY5_SOYBN WD_REPEATS_REGION domain-containing protein OS=Glycine max OX=3847 GN=GLYMA_17G207300 PE=4 SV=1
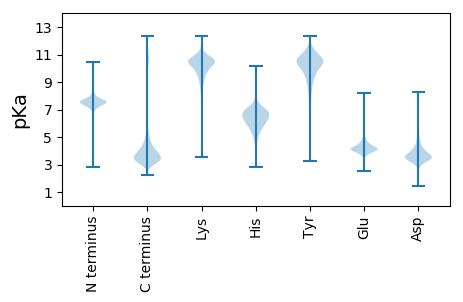
MM1 pKa = 7.76ADD3 pKa = 2.79VTTYY7 pKa = 10.67LRR9 pKa = 11.84HH10 pKa = 6.46HH11 pKa = 7.57PDD13 pKa = 3.09SDD15 pKa = 4.17PNPNPNPNPNPNPDD29 pKa = 4.23DD30 pKa = 4.51DD31 pKa = 3.91QTLIPFPYY39 pKa = 9.11WDD41 pKa = 4.87LDD43 pKa = 3.55FDD45 pKa = 5.29FDD47 pKa = 5.46PEE49 pKa = 4.58FPSNTLSFTDD59 pKa = 4.82RR60 pKa = 11.84EE61 pKa = 4.22NQVNFIMDD69 pKa = 4.73LFHH72 pKa = 7.46QSVEE76 pKa = 4.16QSQLTDD82 pKa = 3.85PLSNDD87 pKa = 3.19AVFGAIDD94 pKa = 4.9GIDD97 pKa = 3.62LGFPAADD104 pKa = 3.82DD105 pKa = 4.09FFVGQRR111 pKa = 11.84FSVGSDD117 pKa = 3.01EE118 pKa = 5.08SHH120 pKa = 5.85THH122 pKa = 5.45PHH124 pKa = 5.6TLAANDD130 pKa = 3.95DD131 pKa = 4.18GVLGFCAHH139 pKa = 6.42SNEE142 pKa = 4.06NDD144 pKa = 3.61DD145 pKa = 3.72VASIPLCWDD154 pKa = 3.83ALQLEE159 pKa = 4.81EE160 pKa = 5.38NNNNNNTYY168 pKa = 11.1EE169 pKa = 4.07DD170 pKa = 4.43FEE172 pKa = 4.57WEE174 pKa = 4.05EE175 pKa = 4.15VMDD178 pKa = 4.11EE179 pKa = 4.17RR180 pKa = 11.84DD181 pKa = 4.3VISMLDD187 pKa = 3.48DD188 pKa = 3.67TVSVSLGIEE197 pKa = 4.2EE198 pKa = 4.11EE199 pKa = 4.5TEE201 pKa = 3.6AAAAEE206 pKa = 4.03EE207 pKa = 4.36DD208 pKa = 3.95AEE210 pKa = 4.65SEE212 pKa = 4.34VSILEE217 pKa = 4.08WQVLLNSTNLEE228 pKa = 4.29GPNSEE233 pKa = 4.82PYY235 pKa = 10.57FGDD238 pKa = 3.42SEE240 pKa = 5.02DD241 pKa = 3.78FVYY244 pKa = 9.68TAEE247 pKa = 4.07YY248 pKa = 11.07EE249 pKa = 4.32MMFGQFNDD257 pKa = 3.1NAFNGKK263 pKa = 8.54PPASASIVRR272 pKa = 11.84SLPSVVVTEE281 pKa = 4.61ADD283 pKa = 3.42VANDD287 pKa = 3.25NNVVVVCAVCKK298 pKa = 10.78DD299 pKa = 3.55EE300 pKa = 4.95FGVGEE305 pKa = 4.3GVKK308 pKa = 10.21VLPCSHH314 pKa = 7.35RR315 pKa = 11.84YY316 pKa = 9.2HH317 pKa = 7.02GEE319 pKa = 4.17CIVPWLGIRR328 pKa = 11.84NTCPVCRR335 pKa = 11.84YY336 pKa = 7.43EE337 pKa = 5.81FPTDD341 pKa = 3.36DD342 pKa = 3.85ADD344 pKa = 3.72YY345 pKa = 10.81EE346 pKa = 4.12RR347 pKa = 11.84RR348 pKa = 11.84KK349 pKa = 9.84AQRR352 pKa = 11.84SVMM355 pKa = 3.78
MM1 pKa = 7.76ADD3 pKa = 2.79VTTYY7 pKa = 10.67LRR9 pKa = 11.84HH10 pKa = 6.46HH11 pKa = 7.57PDD13 pKa = 3.09SDD15 pKa = 4.17PNPNPNPNPNPNPDD29 pKa = 4.23DD30 pKa = 4.51DD31 pKa = 3.91QTLIPFPYY39 pKa = 9.11WDD41 pKa = 4.87LDD43 pKa = 3.55FDD45 pKa = 5.29FDD47 pKa = 5.46PEE49 pKa = 4.58FPSNTLSFTDD59 pKa = 4.82RR60 pKa = 11.84EE61 pKa = 4.22NQVNFIMDD69 pKa = 4.73LFHH72 pKa = 7.46QSVEE76 pKa = 4.16QSQLTDD82 pKa = 3.85PLSNDD87 pKa = 3.19AVFGAIDD94 pKa = 4.9GIDD97 pKa = 3.62LGFPAADD104 pKa = 3.82DD105 pKa = 4.09FFVGQRR111 pKa = 11.84FSVGSDD117 pKa = 3.01EE118 pKa = 5.08SHH120 pKa = 5.85THH122 pKa = 5.45PHH124 pKa = 5.6TLAANDD130 pKa = 3.95DD131 pKa = 4.18GVLGFCAHH139 pKa = 6.42SNEE142 pKa = 4.06NDD144 pKa = 3.61DD145 pKa = 3.72VASIPLCWDD154 pKa = 3.83ALQLEE159 pKa = 4.81EE160 pKa = 5.38NNNNNNTYY168 pKa = 11.1EE169 pKa = 4.07DD170 pKa = 4.43FEE172 pKa = 4.57WEE174 pKa = 4.05EE175 pKa = 4.15VMDD178 pKa = 4.11EE179 pKa = 4.17RR180 pKa = 11.84DD181 pKa = 4.3VISMLDD187 pKa = 3.48DD188 pKa = 3.67TVSVSLGIEE197 pKa = 4.2EE198 pKa = 4.11EE199 pKa = 4.5TEE201 pKa = 3.6AAAAEE206 pKa = 4.03EE207 pKa = 4.36DD208 pKa = 3.95AEE210 pKa = 4.65SEE212 pKa = 4.34VSILEE217 pKa = 4.08WQVLLNSTNLEE228 pKa = 4.29GPNSEE233 pKa = 4.82PYY235 pKa = 10.57FGDD238 pKa = 3.42SEE240 pKa = 5.02DD241 pKa = 3.78FVYY244 pKa = 9.68TAEE247 pKa = 4.07YY248 pKa = 11.07EE249 pKa = 4.32MMFGQFNDD257 pKa = 3.1NAFNGKK263 pKa = 8.54PPASASIVRR272 pKa = 11.84SLPSVVVTEE281 pKa = 4.61ADD283 pKa = 3.42VANDD287 pKa = 3.25NNVVVVCAVCKK298 pKa = 10.78DD299 pKa = 3.55EE300 pKa = 4.95FGVGEE305 pKa = 4.3GVKK308 pKa = 10.21VLPCSHH314 pKa = 7.35RR315 pKa = 11.84YY316 pKa = 9.2HH317 pKa = 7.02GEE319 pKa = 4.17CIVPWLGIRR328 pKa = 11.84NTCPVCRR335 pKa = 11.84YY336 pKa = 7.43EE337 pKa = 5.81FPTDD341 pKa = 3.36DD342 pKa = 3.85ADD344 pKa = 3.72YY345 pKa = 10.81EE346 pKa = 4.12RR347 pKa = 11.84RR348 pKa = 11.84KK349 pKa = 9.84AQRR352 pKa = 11.84SVMM355 pKa = 3.78
Molecular weight: 39.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q0PJI9|Q0PJI9_SOYBN MYB transcription factor MYB124 OS=Glycine max OX=3847 GN=MYB124 PE=2 SV=1
MM1 pKa = 7.38GGVGKK6 pKa = 7.98TKK8 pKa = 10.43RR9 pKa = 11.84MRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 10.24RR15 pKa = 11.84KK16 pKa = 8.21RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.46MRR21 pKa = 11.84QRR23 pKa = 11.84SKK25 pKa = 11.41
MM1 pKa = 7.38GGVGKK6 pKa = 7.98TKK8 pKa = 10.43RR9 pKa = 11.84MRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 10.24RR15 pKa = 11.84KK16 pKa = 8.21RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.46MRR21 pKa = 11.84QRR23 pKa = 11.84SKK25 pKa = 11.41
Molecular weight: 3.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
30697910 |
10 |
5435 |
410.0 |
45.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.487 ± 0.008 | 1.907 ± 0.004 |
5.279 ± 0.006 | 6.387 ± 0.01 |
4.238 ± 0.006 | 6.309 ± 0.008 |
2.55 ± 0.004 | 5.386 ± 0.007 |
6.208 ± 0.009 | 9.788 ± 0.01 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.411 ± 0.003 | 4.739 ± 0.006 |
4.85 ± 0.009 | 3.791 ± 0.007 |
5.108 ± 0.007 | 9.02 ± 0.011 |
4.937 ± 0.005 | 6.502 ± 0.006 |
1.274 ± 0.003 | 2.827 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
