
Natrarchaeobius halalkaliphilus
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Natrarchaeobius
Average proteome isoelectric point is 4.85
Get precalculated fractions of proteins
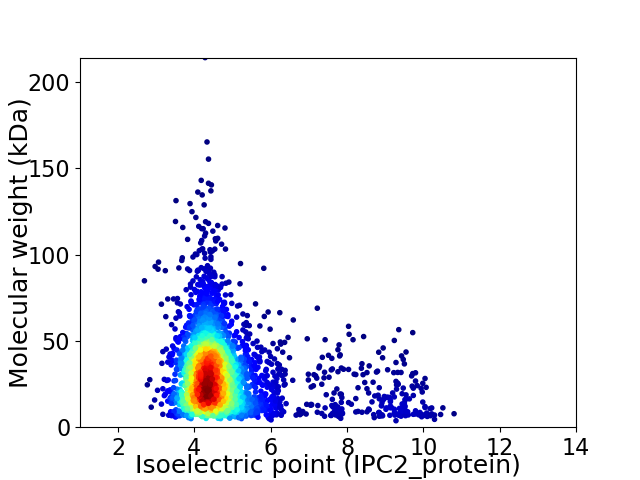
Virtual 2D-PAGE plot for 3330 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N6NWK0|A0A3N6NWK0_9EURY ArsR family transcriptional regulator OS=Natrarchaeobius halalkaliphilus OX=1679091 GN=EA462_11025 PE=4 SV=1
MM1 pKa = 7.41KK2 pKa = 10.4RR3 pKa = 11.84LLLIVLVAAMLVLAGCADD21 pKa = 3.81GAITEE26 pKa = 4.61DD27 pKa = 4.38DD28 pKa = 3.99GTTVEE33 pKa = 4.9ADD35 pKa = 4.31DD36 pKa = 4.41NFEE39 pKa = 4.94DD40 pKa = 5.4ADD42 pKa = 4.72SNDD45 pKa = 3.87ADD47 pKa = 5.76DD48 pKa = 5.76DD49 pKa = 4.21TTTDD53 pKa = 4.02DD54 pKa = 5.78DD55 pKa = 4.01EE56 pKa = 5.19TDD58 pKa = 3.42TEE60 pKa = 4.39EE61 pKa = 4.29TGEE64 pKa = 4.26SEE66 pKa = 5.1AGEE69 pKa = 4.62PDD71 pKa = 3.81PGDD74 pKa = 3.78PDD76 pKa = 3.23VDD78 pKa = 3.87GEE80 pKa = 4.69LEE82 pKa = 3.78IHH84 pKa = 6.89HH85 pKa = 7.0IDD87 pKa = 4.14VGQADD92 pKa = 4.2ATLLIEE98 pKa = 4.52PSGEE102 pKa = 4.13TILIDD107 pKa = 4.32SGDD110 pKa = 3.44WRR112 pKa = 11.84QGGSDD117 pKa = 3.63VIEE120 pKa = 4.0YY121 pKa = 10.79LGDD124 pKa = 3.51QDD126 pKa = 4.63IDD128 pKa = 4.7RR129 pKa = 11.84IDD131 pKa = 3.85HH132 pKa = 6.38LVATHH137 pKa = 5.68GHH139 pKa = 6.58ADD141 pKa = 3.61HH142 pKa = 7.2IGGHH146 pKa = 5.85DD147 pKa = 4.36AIIEE151 pKa = 4.25HH152 pKa = 6.64YY153 pKa = 8.04EE154 pKa = 3.97TEE156 pKa = 3.92RR157 pKa = 11.84DD158 pKa = 4.24GIGVAYY164 pKa = 10.25DD165 pKa = 3.04SGIAHH170 pKa = 7.16TSQTYY175 pKa = 9.16EE176 pKa = 3.99RR177 pKa = 11.84YY178 pKa = 9.62LDD180 pKa = 4.42AIEE183 pKa = 4.12EE184 pKa = 4.31HH185 pKa = 7.31DD186 pKa = 4.08VEE188 pKa = 4.5LLVVEE193 pKa = 4.93EE194 pKa = 4.48GDD196 pKa = 3.51HH197 pKa = 7.81VEE199 pKa = 4.99FGDD202 pKa = 3.87ATVDD206 pKa = 3.61VLNPPAGNSGSDD218 pKa = 2.92LHH220 pKa = 6.85YY221 pKa = 11.17NSVALSIEE229 pKa = 4.09FGEE232 pKa = 4.43FSYY235 pKa = 9.83LTTGDD240 pKa = 3.85AEE242 pKa = 4.62ADD244 pKa = 3.21AEE246 pKa = 4.0QRR248 pKa = 11.84MVDD251 pKa = 3.34EE252 pKa = 4.7HH253 pKa = 7.38GEE255 pKa = 4.04RR256 pKa = 11.84LEE258 pKa = 4.06TDD260 pKa = 3.71AYY262 pKa = 10.19QAGHH266 pKa = 6.66HH267 pKa = 6.49GSTTSSTTPFMDD279 pKa = 2.93QATPEE284 pKa = 4.01VAIISSAYY292 pKa = 9.65DD293 pKa = 3.16SQYY296 pKa = 11.13GHH298 pKa = 6.95PHH300 pKa = 6.9DD301 pKa = 5.27EE302 pKa = 4.19VLEE305 pKa = 4.76DD306 pKa = 3.61YY307 pKa = 10.99ADD309 pKa = 4.12RR310 pKa = 11.84GIEE313 pKa = 4.25TYY315 pKa = 7.55WTAVHH320 pKa = 6.79GDD322 pKa = 3.38VALTTDD328 pKa = 3.47GSDD331 pKa = 3.29VEE333 pKa = 5.54LEE335 pKa = 4.37TEE337 pKa = 4.56HH338 pKa = 6.94EE339 pKa = 4.46FSTDD343 pKa = 3.13AGDD346 pKa = 4.21LLEE349 pKa = 5.37EE350 pKa = 4.89KK351 pKa = 10.27PADD354 pKa = 4.89DD355 pKa = 6.49DD356 pKa = 3.9DD357 pKa = 4.41TQASLTHH364 pKa = 6.94PIDD367 pKa = 3.61VATTPLAGG375 pKa = 3.6
MM1 pKa = 7.41KK2 pKa = 10.4RR3 pKa = 11.84LLLIVLVAAMLVLAGCADD21 pKa = 3.81GAITEE26 pKa = 4.61DD27 pKa = 4.38DD28 pKa = 3.99GTTVEE33 pKa = 4.9ADD35 pKa = 4.31DD36 pKa = 4.41NFEE39 pKa = 4.94DD40 pKa = 5.4ADD42 pKa = 4.72SNDD45 pKa = 3.87ADD47 pKa = 5.76DD48 pKa = 5.76DD49 pKa = 4.21TTTDD53 pKa = 4.02DD54 pKa = 5.78DD55 pKa = 4.01EE56 pKa = 5.19TDD58 pKa = 3.42TEE60 pKa = 4.39EE61 pKa = 4.29TGEE64 pKa = 4.26SEE66 pKa = 5.1AGEE69 pKa = 4.62PDD71 pKa = 3.81PGDD74 pKa = 3.78PDD76 pKa = 3.23VDD78 pKa = 3.87GEE80 pKa = 4.69LEE82 pKa = 3.78IHH84 pKa = 6.89HH85 pKa = 7.0IDD87 pKa = 4.14VGQADD92 pKa = 4.2ATLLIEE98 pKa = 4.52PSGEE102 pKa = 4.13TILIDD107 pKa = 4.32SGDD110 pKa = 3.44WRR112 pKa = 11.84QGGSDD117 pKa = 3.63VIEE120 pKa = 4.0YY121 pKa = 10.79LGDD124 pKa = 3.51QDD126 pKa = 4.63IDD128 pKa = 4.7RR129 pKa = 11.84IDD131 pKa = 3.85HH132 pKa = 6.38LVATHH137 pKa = 5.68GHH139 pKa = 6.58ADD141 pKa = 3.61HH142 pKa = 7.2IGGHH146 pKa = 5.85DD147 pKa = 4.36AIIEE151 pKa = 4.25HH152 pKa = 6.64YY153 pKa = 8.04EE154 pKa = 3.97TEE156 pKa = 3.92RR157 pKa = 11.84DD158 pKa = 4.24GIGVAYY164 pKa = 10.25DD165 pKa = 3.04SGIAHH170 pKa = 7.16TSQTYY175 pKa = 9.16EE176 pKa = 3.99RR177 pKa = 11.84YY178 pKa = 9.62LDD180 pKa = 4.42AIEE183 pKa = 4.12EE184 pKa = 4.31HH185 pKa = 7.31DD186 pKa = 4.08VEE188 pKa = 4.5LLVVEE193 pKa = 4.93EE194 pKa = 4.48GDD196 pKa = 3.51HH197 pKa = 7.81VEE199 pKa = 4.99FGDD202 pKa = 3.87ATVDD206 pKa = 3.61VLNPPAGNSGSDD218 pKa = 2.92LHH220 pKa = 6.85YY221 pKa = 11.17NSVALSIEE229 pKa = 4.09FGEE232 pKa = 4.43FSYY235 pKa = 9.83LTTGDD240 pKa = 3.85AEE242 pKa = 4.62ADD244 pKa = 3.21AEE246 pKa = 4.0QRR248 pKa = 11.84MVDD251 pKa = 3.34EE252 pKa = 4.7HH253 pKa = 7.38GEE255 pKa = 4.04RR256 pKa = 11.84LEE258 pKa = 4.06TDD260 pKa = 3.71AYY262 pKa = 10.19QAGHH266 pKa = 6.66HH267 pKa = 6.49GSTTSSTTPFMDD279 pKa = 2.93QATPEE284 pKa = 4.01VAIISSAYY292 pKa = 9.65DD293 pKa = 3.16SQYY296 pKa = 11.13GHH298 pKa = 6.95PHH300 pKa = 6.9DD301 pKa = 5.27EE302 pKa = 4.19VLEE305 pKa = 4.76DD306 pKa = 3.61YY307 pKa = 10.99ADD309 pKa = 4.12RR310 pKa = 11.84GIEE313 pKa = 4.25TYY315 pKa = 7.55WTAVHH320 pKa = 6.79GDD322 pKa = 3.38VALTTDD328 pKa = 3.47GSDD331 pKa = 3.29VEE333 pKa = 5.54LEE335 pKa = 4.37TEE337 pKa = 4.56HH338 pKa = 6.94EE339 pKa = 4.46FSTDD343 pKa = 3.13AGDD346 pKa = 4.21LLEE349 pKa = 5.37EE350 pKa = 4.89KK351 pKa = 10.27PADD354 pKa = 4.89DD355 pKa = 6.49DD356 pKa = 3.9DD357 pKa = 4.41TQASLTHH364 pKa = 6.94PIDD367 pKa = 3.61VATTPLAGG375 pKa = 3.6
Molecular weight: 40.33 kDa
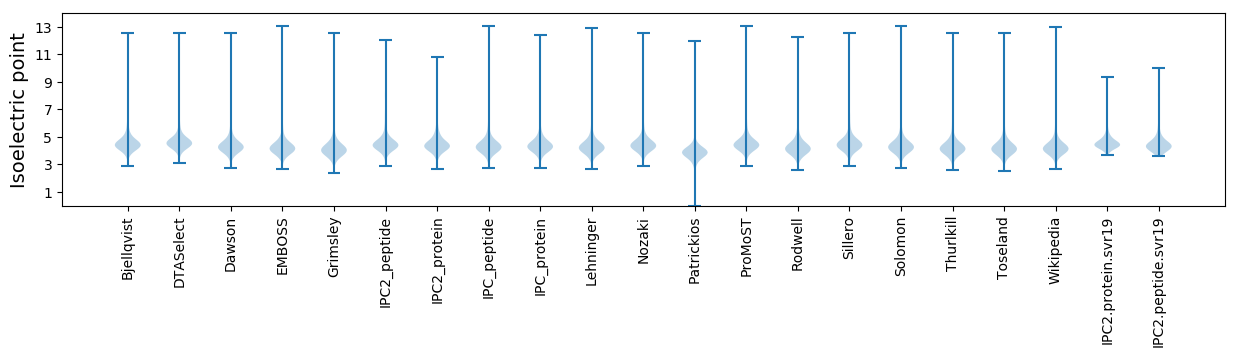
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N6LQ48|A0A3N6LQ48_9EURY Acyl-CoA carboxylase subunit beta OS=Natrarchaeobius halalkaliphilus OX=1679091 GN=EA462_03600 PE=4 SV=1
MM1 pKa = 7.14VLQRR5 pKa = 11.84FLGSKK10 pKa = 8.46ATRR13 pKa = 11.84SLTVVSVLMEE23 pKa = 4.67AKK25 pKa = 10.07RR26 pKa = 11.84ALARR30 pKa = 11.84GRR32 pKa = 11.84RR33 pKa = 11.84TRR35 pKa = 11.84GLVLLGVAVVAWKK48 pKa = 8.04WTVIGLAAQGVIKK61 pKa = 10.39ALRR64 pKa = 11.84AGRR67 pKa = 11.84PSGSSSAA74 pKa = 3.79
MM1 pKa = 7.14VLQRR5 pKa = 11.84FLGSKK10 pKa = 8.46ATRR13 pKa = 11.84SLTVVSVLMEE23 pKa = 4.67AKK25 pKa = 10.07RR26 pKa = 11.84ALARR30 pKa = 11.84GRR32 pKa = 11.84RR33 pKa = 11.84TRR35 pKa = 11.84GLVLLGVAVVAWKK48 pKa = 8.04WTVIGLAAQGVIKK61 pKa = 10.39ALRR64 pKa = 11.84AGRR67 pKa = 11.84PSGSSSAA74 pKa = 3.79
Molecular weight: 7.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
976133 |
32 |
1921 |
293.1 |
32.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.759 ± 0.057 | 0.761 ± 0.014 |
8.587 ± 0.066 | 9.119 ± 0.057 |
3.27 ± 0.031 | 8.196 ± 0.04 |
2.053 ± 0.018 | 4.775 ± 0.034 |
1.821 ± 0.025 | 8.727 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.75 ± 0.021 | 2.456 ± 0.022 |
4.525 ± 0.024 | 2.383 ± 0.024 |
6.686 ± 0.049 | 6.037 ± 0.035 |
6.548 ± 0.031 | 8.76 ± 0.042 |
1.085 ± 0.017 | 2.702 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
