
Monosporascus sp. MG133
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Xylariomycetidae; Xylariales; Xylariales incertae sedis; Monosporascus; unclassified Monosporascus
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
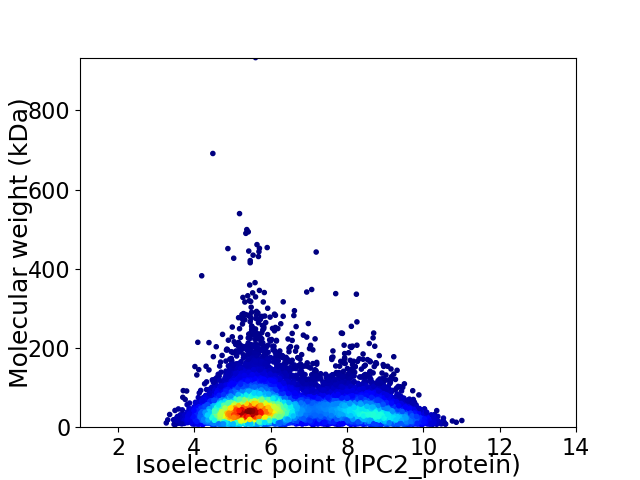
Virtual 2D-PAGE plot for 11566 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q4V6C8|A0A4Q4V6C8_9PEZI Uncharacterized protein OS=Monosporascus sp. MG133 OX=2211645 GN=DL767_006947 PE=4 SV=1
MM1 pKa = 7.99RR2 pKa = 11.84KK3 pKa = 8.75PLSVSKK9 pKa = 10.31RR10 pKa = 11.84QEE12 pKa = 3.88DD13 pKa = 3.86TGVPLHH19 pKa = 7.18DD20 pKa = 4.08ISQVSYY26 pKa = 10.84LIEE29 pKa = 4.13LSIGTPGQSVKK40 pKa = 10.7VAVDD44 pKa = 3.52TGSSEE49 pKa = 4.32LWVDD53 pKa = 4.05PVCQTSQSLSEE64 pKa = 4.27AEE66 pKa = 3.79EE67 pKa = 4.75CIANGIYY74 pKa = 10.38DD75 pKa = 4.29PSRR78 pKa = 11.84SDD80 pKa = 3.04TFEE83 pKa = 4.92DD84 pKa = 3.92LNSTNTIPYY93 pKa = 9.95GIGAVQIEE101 pKa = 4.57YY102 pKa = 10.79VSDD105 pKa = 4.15NIAFPDD111 pKa = 3.58STINLTDD118 pKa = 3.28VQFGVATASRR128 pKa = 11.84QLNEE132 pKa = 4.58GIMGLSFGGNTPEE145 pKa = 5.82ADD147 pKa = 3.15LSYY150 pKa = 11.6NNIVDD155 pKa = 4.0EE156 pKa = 5.73LYY158 pKa = 10.8LQNATQSRR166 pKa = 11.84AFSVALGSSDD176 pKa = 3.65SDD178 pKa = 3.51NDD180 pKa = 3.6SVLIFGGIDD189 pKa = 3.1TSKK192 pKa = 10.49YY193 pKa = 10.15AGPLRR198 pKa = 11.84TLPILGPQNGEE209 pKa = 3.61TIYY212 pKa = 10.5RR213 pKa = 11.84YY214 pKa = 8.57WVQLDD219 pKa = 4.07SIGATVDD226 pKa = 3.99GSSHH230 pKa = 6.46TYY232 pKa = 10.38DD233 pKa = 3.31SSSLPVVLDD242 pKa = 3.52TGSTFCSLPRR252 pKa = 11.84AVVNGMMEE260 pKa = 4.5DD261 pKa = 3.78LDD263 pKa = 4.16GQIDD267 pKa = 3.67RR268 pKa = 11.84QGQVLVDD275 pKa = 3.87CSQVDD280 pKa = 3.54NEE282 pKa = 4.7DD283 pKa = 3.38TFDD286 pKa = 4.12FDD288 pKa = 4.88FGNITIRR295 pKa = 11.84IPYY298 pKa = 8.82SQFVVQANSEE308 pKa = 4.45VCVLGAIPDD317 pKa = 3.86DD318 pKa = 4.26QIALLGDD325 pKa = 3.49SFLRR329 pKa = 11.84SAYY332 pKa = 10.54VVFDD336 pKa = 3.8QNNMEE341 pKa = 4.5IAMAPYY347 pKa = 10.05ANCGQSEE354 pKa = 3.96QRR356 pKa = 11.84IPDD359 pKa = 3.52EE360 pKa = 4.36GVSGVEE366 pKa = 4.34GACDD370 pKa = 3.28SANNDD375 pKa = 3.22GGGDD379 pKa = 3.55SGGGDD384 pKa = 3.85DD385 pKa = 4.6NAGSRR390 pKa = 11.84LTASMLGLLSACISVPVVVDD410 pKa = 3.78LLL412 pKa = 3.95
MM1 pKa = 7.99RR2 pKa = 11.84KK3 pKa = 8.75PLSVSKK9 pKa = 10.31RR10 pKa = 11.84QEE12 pKa = 3.88DD13 pKa = 3.86TGVPLHH19 pKa = 7.18DD20 pKa = 4.08ISQVSYY26 pKa = 10.84LIEE29 pKa = 4.13LSIGTPGQSVKK40 pKa = 10.7VAVDD44 pKa = 3.52TGSSEE49 pKa = 4.32LWVDD53 pKa = 4.05PVCQTSQSLSEE64 pKa = 4.27AEE66 pKa = 3.79EE67 pKa = 4.75CIANGIYY74 pKa = 10.38DD75 pKa = 4.29PSRR78 pKa = 11.84SDD80 pKa = 3.04TFEE83 pKa = 4.92DD84 pKa = 3.92LNSTNTIPYY93 pKa = 9.95GIGAVQIEE101 pKa = 4.57YY102 pKa = 10.79VSDD105 pKa = 4.15NIAFPDD111 pKa = 3.58STINLTDD118 pKa = 3.28VQFGVATASRR128 pKa = 11.84QLNEE132 pKa = 4.58GIMGLSFGGNTPEE145 pKa = 5.82ADD147 pKa = 3.15LSYY150 pKa = 11.6NNIVDD155 pKa = 4.0EE156 pKa = 5.73LYY158 pKa = 10.8LQNATQSRR166 pKa = 11.84AFSVALGSSDD176 pKa = 3.65SDD178 pKa = 3.51NDD180 pKa = 3.6SVLIFGGIDD189 pKa = 3.1TSKK192 pKa = 10.49YY193 pKa = 10.15AGPLRR198 pKa = 11.84TLPILGPQNGEE209 pKa = 3.61TIYY212 pKa = 10.5RR213 pKa = 11.84YY214 pKa = 8.57WVQLDD219 pKa = 4.07SIGATVDD226 pKa = 3.99GSSHH230 pKa = 6.46TYY232 pKa = 10.38DD233 pKa = 3.31SSSLPVVLDD242 pKa = 3.52TGSTFCSLPRR252 pKa = 11.84AVVNGMMEE260 pKa = 4.5DD261 pKa = 3.78LDD263 pKa = 4.16GQIDD267 pKa = 3.67RR268 pKa = 11.84QGQVLVDD275 pKa = 3.87CSQVDD280 pKa = 3.54NEE282 pKa = 4.7DD283 pKa = 3.38TFDD286 pKa = 4.12FDD288 pKa = 4.88FGNITIRR295 pKa = 11.84IPYY298 pKa = 8.82SQFVVQANSEE308 pKa = 4.45VCVLGAIPDD317 pKa = 3.86DD318 pKa = 4.26QIALLGDD325 pKa = 3.49SFLRR329 pKa = 11.84SAYY332 pKa = 10.54VVFDD336 pKa = 3.8QNNMEE341 pKa = 4.5IAMAPYY347 pKa = 10.05ANCGQSEE354 pKa = 3.96QRR356 pKa = 11.84IPDD359 pKa = 3.52EE360 pKa = 4.36GVSGVEE366 pKa = 4.34GACDD370 pKa = 3.28SANNDD375 pKa = 3.22GGGDD379 pKa = 3.55SGGGDD384 pKa = 3.85DD385 pKa = 4.6NAGSRR390 pKa = 11.84LTASMLGLLSACISVPVVVDD410 pKa = 3.78LLL412 pKa = 3.95
Molecular weight: 43.75 kDa
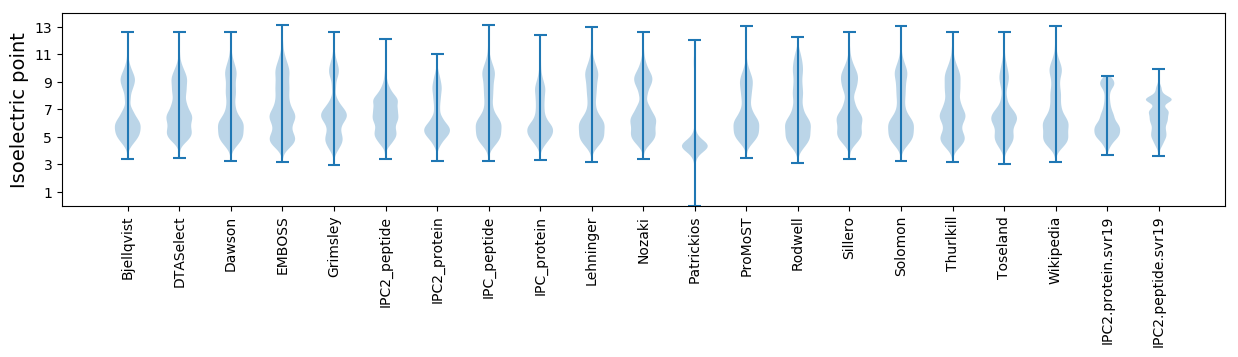
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q4U5B3|A0A4Q4U5B3_9PEZI MICOS complex subunit MIC60 OS=Monosporascus sp. MG133 OX=2211645 GN=DL767_010748 PE=3 SV=1
MM1 pKa = 7.62AKK3 pKa = 10.3ASSTVTTGRR12 pKa = 11.84TVTAGRR18 pKa = 11.84MATAHH23 pKa = 6.52HH24 pKa = 6.79GPPGRR29 pKa = 11.84HH30 pKa = 5.17MKK32 pKa = 10.79LPIQVAFAMGRR43 pKa = 11.84TKK45 pKa = 10.6VGSKK49 pKa = 11.01AMVDD53 pKa = 3.2NKK55 pKa = 9.8VRR57 pKa = 11.84AEE59 pKa = 3.79NRR61 pKa = 11.84AKK63 pKa = 10.4GRR65 pKa = 11.84SRR67 pKa = 11.84ARR69 pKa = 11.84TSHH72 pKa = 6.59LPMVRR77 pKa = 11.84RR78 pKa = 11.84LWWSLAPQGLRR89 pKa = 11.84LRR91 pKa = 11.84LLSKK95 pKa = 10.36PLAPSPITATTVVTMTRR112 pKa = 11.84RR113 pKa = 11.84TT114 pKa = 3.46
MM1 pKa = 7.62AKK3 pKa = 10.3ASSTVTTGRR12 pKa = 11.84TVTAGRR18 pKa = 11.84MATAHH23 pKa = 6.52HH24 pKa = 6.79GPPGRR29 pKa = 11.84HH30 pKa = 5.17MKK32 pKa = 10.79LPIQVAFAMGRR43 pKa = 11.84TKK45 pKa = 10.6VGSKK49 pKa = 11.01AMVDD53 pKa = 3.2NKK55 pKa = 9.8VRR57 pKa = 11.84AEE59 pKa = 3.79NRR61 pKa = 11.84AKK63 pKa = 10.4GRR65 pKa = 11.84SRR67 pKa = 11.84ARR69 pKa = 11.84TSHH72 pKa = 6.59LPMVRR77 pKa = 11.84RR78 pKa = 11.84LWWSLAPQGLRR89 pKa = 11.84LRR91 pKa = 11.84LLSKK95 pKa = 10.36PLAPSPITATTVVTMTRR112 pKa = 11.84RR113 pKa = 11.84TT114 pKa = 3.46
Molecular weight: 12.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5736655 |
66 |
8424 |
496.0 |
54.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.248 ± 0.023 | 1.177 ± 0.008 |
5.912 ± 0.017 | 6.342 ± 0.021 |
3.543 ± 0.015 | 7.305 ± 0.022 |
2.341 ± 0.009 | 4.514 ± 0.015 |
4.589 ± 0.021 | 8.759 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.105 ± 0.009 | 3.505 ± 0.013 |
6.213 ± 0.031 | 3.856 ± 0.015 |
6.584 ± 0.019 | 7.835 ± 0.023 |
5.778 ± 0.015 | 6.212 ± 0.017 |
1.47 ± 0.009 | 2.712 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
