
Labidocera aestiva circovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 8.74
Get precalculated fractions of proteins
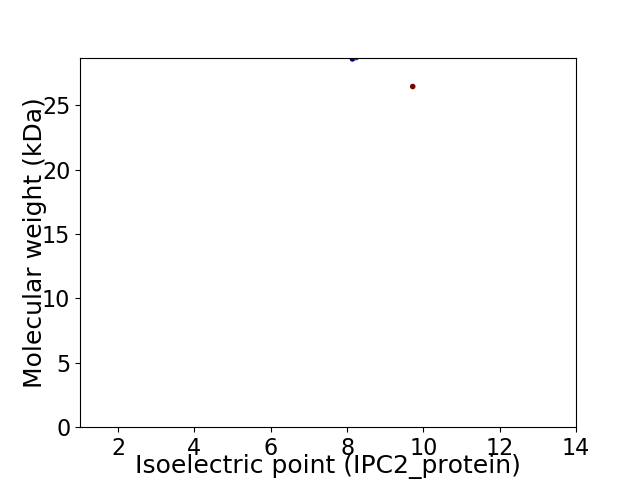
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
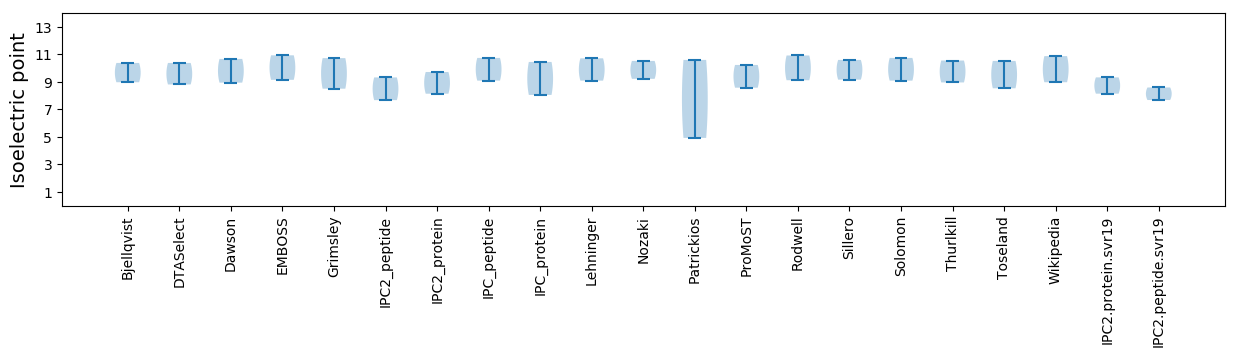
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M3VMK9|M3VMK9_9CIRC Putative viral replication protein OS=Labidocera aestiva circovirus OX=1173709 GN=LaCopCV_gp1 PE=4 SV=1
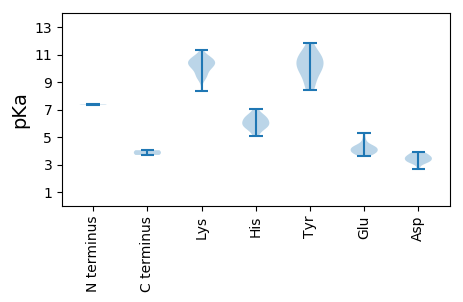
MM1 pKa = 7.38SRR3 pKa = 11.84NFTGTVNNPTQTLEE17 pKa = 3.69EE18 pKa = 4.08WLATLKK24 pKa = 10.64RR25 pKa = 11.84LPHH28 pKa = 5.64ATAARR33 pKa = 11.84CQLEE37 pKa = 4.11KK38 pKa = 11.31GEE40 pKa = 4.66NGTPHH45 pKa = 6.58IQWCVTLSKK54 pKa = 10.45VARR57 pKa = 11.84LTGIIKK63 pKa = 10.3KK64 pKa = 10.05LAGSHH69 pKa = 5.79VEE71 pKa = 3.8ASRR74 pKa = 11.84NAMAAWKK81 pKa = 10.25YY82 pKa = 10.17CGKK85 pKa = 10.26EE86 pKa = 3.87DD87 pKa = 3.86TRR89 pKa = 11.84VEE91 pKa = 4.31GPLEE95 pKa = 4.35FGVPPASKK103 pKa = 9.9RR104 pKa = 11.84VKK106 pKa = 10.72GDD108 pKa = 3.14TKK110 pKa = 10.74EE111 pKa = 3.94RR112 pKa = 11.84NKK114 pKa = 10.23MILEE118 pKa = 4.91LGPVAAVEE126 pKa = 3.93QGLIPVEE133 pKa = 3.82KK134 pKa = 10.26FKK136 pKa = 10.93AIKK139 pKa = 9.86QSCDD143 pKa = 2.7LFAVMKK149 pKa = 10.61KK150 pKa = 9.24EE151 pKa = 3.88VHH153 pKa = 5.35TTDD156 pKa = 3.27ALSNEE161 pKa = 3.64WHH163 pKa = 6.36YY164 pKa = 11.81GEE166 pKa = 5.27SGTGKK171 pKa = 9.89SRR173 pKa = 11.84GVRR176 pKa = 11.84EE177 pKa = 4.16RR178 pKa = 11.84FPDD181 pKa = 3.63AFIKK185 pKa = 10.93SSDD188 pKa = 3.68VWWDD192 pKa = 3.88GYY194 pKa = 10.79QGEE197 pKa = 4.36EE198 pKa = 3.89TVIIEE203 pKa = 4.32EE204 pKa = 4.52MGPKK208 pKa = 9.96QIGAHH213 pKa = 5.74HH214 pKa = 6.52LKK216 pKa = 9.61IWSDD220 pKa = 3.47HH221 pKa = 5.78YY222 pKa = 10.88PFKK225 pKa = 10.71AAQKK229 pKa = 10.32GSQLLIRR236 pKa = 11.84PKK238 pKa = 10.44RR239 pKa = 11.84IIVTSNYY246 pKa = 9.94SIKK249 pKa = 10.41EE250 pKa = 4.15CYY252 pKa = 8.72EE253 pKa = 4.01AEE255 pKa = 4.04
MM1 pKa = 7.38SRR3 pKa = 11.84NFTGTVNNPTQTLEE17 pKa = 3.69EE18 pKa = 4.08WLATLKK24 pKa = 10.64RR25 pKa = 11.84LPHH28 pKa = 5.64ATAARR33 pKa = 11.84CQLEE37 pKa = 4.11KK38 pKa = 11.31GEE40 pKa = 4.66NGTPHH45 pKa = 6.58IQWCVTLSKK54 pKa = 10.45VARR57 pKa = 11.84LTGIIKK63 pKa = 10.3KK64 pKa = 10.05LAGSHH69 pKa = 5.79VEE71 pKa = 3.8ASRR74 pKa = 11.84NAMAAWKK81 pKa = 10.25YY82 pKa = 10.17CGKK85 pKa = 10.26EE86 pKa = 3.87DD87 pKa = 3.86TRR89 pKa = 11.84VEE91 pKa = 4.31GPLEE95 pKa = 4.35FGVPPASKK103 pKa = 9.9RR104 pKa = 11.84VKK106 pKa = 10.72GDD108 pKa = 3.14TKK110 pKa = 10.74EE111 pKa = 3.94RR112 pKa = 11.84NKK114 pKa = 10.23MILEE118 pKa = 4.91LGPVAAVEE126 pKa = 3.93QGLIPVEE133 pKa = 3.82KK134 pKa = 10.26FKK136 pKa = 10.93AIKK139 pKa = 9.86QSCDD143 pKa = 2.7LFAVMKK149 pKa = 10.61KK150 pKa = 9.24EE151 pKa = 3.88VHH153 pKa = 5.35TTDD156 pKa = 3.27ALSNEE161 pKa = 3.64WHH163 pKa = 6.36YY164 pKa = 11.81GEE166 pKa = 5.27SGTGKK171 pKa = 9.89SRR173 pKa = 11.84GVRR176 pKa = 11.84EE177 pKa = 4.16RR178 pKa = 11.84FPDD181 pKa = 3.63AFIKK185 pKa = 10.93SSDD188 pKa = 3.68VWWDD192 pKa = 3.88GYY194 pKa = 10.79QGEE197 pKa = 4.36EE198 pKa = 3.89TVIIEE203 pKa = 4.32EE204 pKa = 4.52MGPKK208 pKa = 9.96QIGAHH213 pKa = 5.74HH214 pKa = 6.52LKK216 pKa = 9.61IWSDD220 pKa = 3.47HH221 pKa = 5.78YY222 pKa = 10.88PFKK225 pKa = 10.71AAQKK229 pKa = 10.32GSQLLIRR236 pKa = 11.84PKK238 pKa = 10.44RR239 pKa = 11.84IIVTSNYY246 pKa = 9.94SIKK249 pKa = 10.41EE250 pKa = 4.15CYY252 pKa = 8.72EE253 pKa = 4.01AEE255 pKa = 4.04
Molecular weight: 28.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M3VMK9|M3VMK9_9CIRC Putative viral replication protein OS=Labidocera aestiva circovirus OX=1173709 GN=LaCopCV_gp1 PE=4 SV=1
MM1 pKa = 7.34PARR4 pKa = 11.84RR5 pKa = 11.84SFRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.13SAPKK15 pKa = 9.41RR16 pKa = 11.84RR17 pKa = 11.84ARR19 pKa = 11.84GRR21 pKa = 11.84KK22 pKa = 8.32ARR24 pKa = 11.84ARR26 pKa = 11.84RR27 pKa = 11.84SPFNSSWYY35 pKa = 9.29HH36 pKa = 6.12AKK38 pKa = 10.53CQVTFPIEE46 pKa = 4.05HH47 pKa = 6.6NNNYY51 pKa = 9.9AQGSDD56 pKa = 3.45TTCARR61 pKa = 11.84FVVKK65 pKa = 10.33WNDD68 pKa = 3.08SSNDD72 pKa = 3.47DD73 pKa = 3.38MATPQVAVLRR83 pKa = 11.84NSRR86 pKa = 11.84QWQALAAQFRR96 pKa = 11.84EE97 pKa = 4.22YY98 pKa = 10.17TVRR101 pKa = 11.84GVKK104 pKa = 9.48IEE106 pKa = 4.2YY107 pKa = 9.46KK108 pKa = 10.14GHH110 pKa = 6.07NAVGGDD116 pKa = 3.21KK117 pKa = 10.08EE118 pKa = 4.33YY119 pKa = 10.58RR120 pKa = 11.84ASHH123 pKa = 5.71VWSDD127 pKa = 3.23PDD129 pKa = 3.87VYY131 pKa = 11.01VAPTEE136 pKa = 4.22AGNDD140 pKa = 3.56GAYY143 pKa = 10.3FWDD146 pKa = 3.36INRR149 pKa = 11.84FIQKK153 pKa = 9.42PDD155 pKa = 3.58YY156 pKa = 11.14KK157 pKa = 10.76EE158 pKa = 3.83MSHH161 pKa = 6.11NRR163 pKa = 11.84SFTKK167 pKa = 10.73YY168 pKa = 8.45IGVEE172 pKa = 3.66KK173 pKa = 9.49FHH175 pKa = 7.07RR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84GLNHH182 pKa = 5.06THH184 pKa = 6.18RR185 pKa = 11.84TLAQISTPHH194 pKa = 5.96CTSFQLNSRR203 pKa = 11.84GFDD206 pKa = 3.19NGHH209 pKa = 6.42DD210 pKa = 3.69MGRR213 pKa = 11.84VTVTWFVIFKK223 pKa = 10.58QPAFF227 pKa = 3.7
MM1 pKa = 7.34PARR4 pKa = 11.84RR5 pKa = 11.84SFRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.13SAPKK15 pKa = 9.41RR16 pKa = 11.84RR17 pKa = 11.84ARR19 pKa = 11.84GRR21 pKa = 11.84KK22 pKa = 8.32ARR24 pKa = 11.84ARR26 pKa = 11.84RR27 pKa = 11.84SPFNSSWYY35 pKa = 9.29HH36 pKa = 6.12AKK38 pKa = 10.53CQVTFPIEE46 pKa = 4.05HH47 pKa = 6.6NNNYY51 pKa = 9.9AQGSDD56 pKa = 3.45TTCARR61 pKa = 11.84FVVKK65 pKa = 10.33WNDD68 pKa = 3.08SSNDD72 pKa = 3.47DD73 pKa = 3.38MATPQVAVLRR83 pKa = 11.84NSRR86 pKa = 11.84QWQALAAQFRR96 pKa = 11.84EE97 pKa = 4.22YY98 pKa = 10.17TVRR101 pKa = 11.84GVKK104 pKa = 9.48IEE106 pKa = 4.2YY107 pKa = 9.46KK108 pKa = 10.14GHH110 pKa = 6.07NAVGGDD116 pKa = 3.21KK117 pKa = 10.08EE118 pKa = 4.33YY119 pKa = 10.58RR120 pKa = 11.84ASHH123 pKa = 5.71VWSDD127 pKa = 3.23PDD129 pKa = 3.87VYY131 pKa = 11.01VAPTEE136 pKa = 4.22AGNDD140 pKa = 3.56GAYY143 pKa = 10.3FWDD146 pKa = 3.36INRR149 pKa = 11.84FIQKK153 pKa = 9.42PDD155 pKa = 3.58YY156 pKa = 11.14KK157 pKa = 10.76EE158 pKa = 3.83MSHH161 pKa = 6.11NRR163 pKa = 11.84SFTKK167 pKa = 10.73YY168 pKa = 8.45IGVEE172 pKa = 3.66KK173 pKa = 9.49FHH175 pKa = 7.07RR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84GLNHH182 pKa = 5.06THH184 pKa = 6.18RR185 pKa = 11.84TLAQISTPHH194 pKa = 5.96CTSFQLNSRR203 pKa = 11.84GFDD206 pKa = 3.19NGHH209 pKa = 6.42DD210 pKa = 3.69MGRR213 pKa = 11.84VTVTWFVIFKK223 pKa = 10.58QPAFF227 pKa = 3.7
Molecular weight: 26.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
482 |
227 |
255 |
241.0 |
27.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.506 ± 0.18 | 1.66 ± 0.2 |
4.149 ± 0.672 | 6.224 ± 1.857 |
4.357 ± 1.071 | 6.846 ± 0.662 |
3.734 ± 0.397 | 4.564 ± 0.875 |
7.676 ± 1.153 | 4.564 ± 1.396 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.867 ± 0.062 | 4.564 ± 0.948 |
4.564 ± 0.094 | 3.942 ± 0.274 |
8.091 ± 1.988 | 6.432 ± 0.365 |
6.017 ± 0.171 | 6.432 ± 0.104 |
2.697 ± 0.032 | 3.112 ± 0.504 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
