
Clunio marinus
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera; Nematocera; Culicomorpha; Chironomoidea; Chironomidae; Orthocladiinae; Cl
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
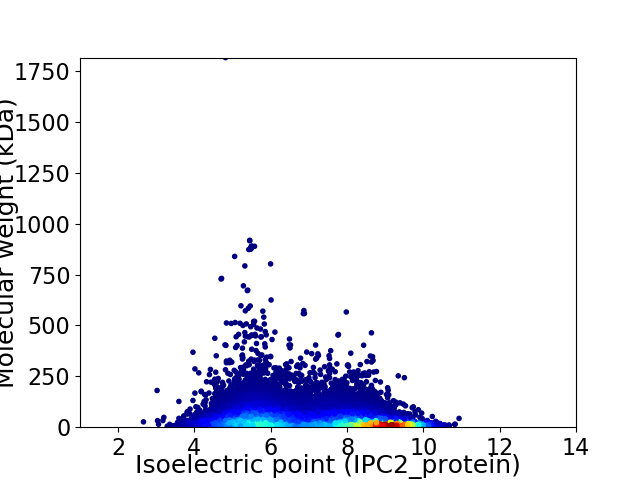
Virtual 2D-PAGE plot for 22544 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1J1I037|A0A1J1I037_9DIPT CLUMA_CG005793 isoform A OS=Clunio marinus OX=568069 GN=CLUMA_CG005793 PE=4 SV=1
MM1 pKa = 7.52KK2 pKa = 10.27LLHH5 pKa = 6.65LFLVVIIAEE14 pKa = 4.34EE15 pKa = 4.08AFSGDD20 pKa = 3.86GNSCSSPPLSFSPSDD35 pKa = 3.36IINRR39 pKa = 11.84LDD41 pKa = 3.83NIKK44 pKa = 10.86DD45 pKa = 4.15FIGDD49 pKa = 3.59AVSEE53 pKa = 4.0YY54 pKa = 11.29LPFFEE59 pKa = 5.71GADD62 pKa = 3.4VDD64 pKa = 4.81FGGTNHH70 pKa = 6.49EE71 pKa = 4.26KK72 pKa = 10.77SEE74 pKa = 4.23NAVSLLIEE82 pKa = 4.34LQTVLSNNGIPSSVQDD98 pKa = 4.41FFPLIDD104 pKa = 4.93DD105 pKa = 5.18IIYY108 pKa = 7.51NTQSTGIQKK117 pKa = 10.64KK118 pKa = 8.63IDD120 pKa = 4.04EE121 pKa = 5.25IIASVDD127 pKa = 3.02AASQILYY134 pKa = 10.32AVLGTIIGQIATEE147 pKa = 4.39TNSSLDD153 pKa = 3.6YY154 pKa = 10.94LSDD157 pKa = 3.9VMTQQIDD164 pKa = 3.55TADD167 pKa = 3.73YY168 pKa = 9.23PISCIAEE175 pKa = 3.97ALIQLYY181 pKa = 9.37QFMFNTYY188 pKa = 7.38EE189 pKa = 4.17TIEE192 pKa = 4.15NAAIAAKK199 pKa = 10.51ASMEE203 pKa = 3.91AMPP206 pKa = 5.42
MM1 pKa = 7.52KK2 pKa = 10.27LLHH5 pKa = 6.65LFLVVIIAEE14 pKa = 4.34EE15 pKa = 4.08AFSGDD20 pKa = 3.86GNSCSSPPLSFSPSDD35 pKa = 3.36IINRR39 pKa = 11.84LDD41 pKa = 3.83NIKK44 pKa = 10.86DD45 pKa = 4.15FIGDD49 pKa = 3.59AVSEE53 pKa = 4.0YY54 pKa = 11.29LPFFEE59 pKa = 5.71GADD62 pKa = 3.4VDD64 pKa = 4.81FGGTNHH70 pKa = 6.49EE71 pKa = 4.26KK72 pKa = 10.77SEE74 pKa = 4.23NAVSLLIEE82 pKa = 4.34LQTVLSNNGIPSSVQDD98 pKa = 4.41FFPLIDD104 pKa = 4.93DD105 pKa = 5.18IIYY108 pKa = 7.51NTQSTGIQKK117 pKa = 10.64KK118 pKa = 8.63IDD120 pKa = 4.04EE121 pKa = 5.25IIASVDD127 pKa = 3.02AASQILYY134 pKa = 10.32AVLGTIIGQIATEE147 pKa = 4.39TNSSLDD153 pKa = 3.6YY154 pKa = 10.94LSDD157 pKa = 3.9VMTQQIDD164 pKa = 3.55TADD167 pKa = 3.73YY168 pKa = 9.23PISCIAEE175 pKa = 3.97ALIQLYY181 pKa = 9.37QFMFNTYY188 pKa = 7.38EE189 pKa = 4.17TIEE192 pKa = 4.15NAAIAAKK199 pKa = 10.51ASMEE203 pKa = 3.91AMPP206 pKa = 5.42
Molecular weight: 22.39 kDa
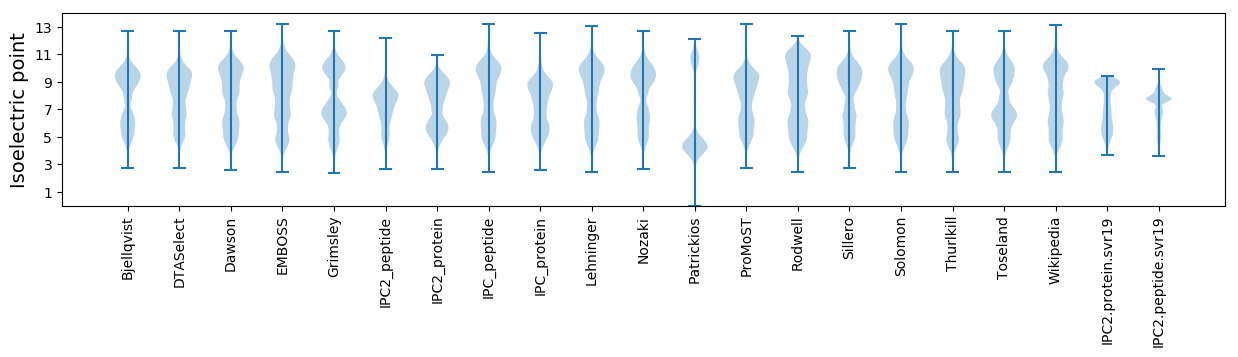
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1J1IDF5|A0A1J1IDF5_9DIPT CLUMA_CG010414 isoform A OS=Clunio marinus OX=568069 GN=CLUMA_CG010414 PE=4 SV=1
MM1 pKa = 6.72NTSTRR6 pKa = 11.84LTLNDD11 pKa = 3.74RR12 pKa = 11.84FSMIKK17 pKa = 9.67QGNGVLPKK25 pKa = 8.84QQRR28 pKa = 11.84GRR30 pKa = 11.84SRR32 pKa = 11.84SRR34 pKa = 11.84SQTRR38 pKa = 11.84RR39 pKa = 11.84PSRR42 pKa = 11.84QLSQGGPRR50 pKa = 11.84KK51 pKa = 9.6NRR53 pKa = 11.84QLLDD57 pKa = 3.52QLEE60 pKa = 4.6KK61 pKa = 8.35QHH63 pKa = 6.03KK64 pKa = 8.75MRR66 pKa = 11.84MAMKK70 pKa = 10.05LKK72 pKa = 9.92NKK74 pKa = 10.23SIMRR78 pKa = 11.84SRR80 pKa = 11.84GRR82 pKa = 11.84VQSQNRR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84PANSLIATIKK100 pKa = 10.41RR101 pKa = 11.84KK102 pKa = 8.28MVEE105 pKa = 4.11DD106 pKa = 3.42SLKK109 pKa = 10.67RR110 pKa = 11.84GSNLTTFVPLNDD122 pKa = 3.51GPRR125 pKa = 11.84RR126 pKa = 11.84NFRR129 pKa = 11.84GRR131 pKa = 11.84SRR133 pKa = 11.84SRR135 pKa = 11.84SQSRR139 pKa = 11.84NRR141 pKa = 11.84NQQRR145 pKa = 11.84SQSRR149 pKa = 11.84NRR151 pKa = 11.84NQQQQQGSVIDD162 pKa = 3.83RR163 pKa = 11.84LGVRR167 pKa = 11.84QPLKK171 pKa = 10.34RR172 pKa = 11.84MRR174 pKa = 11.84RR175 pKa = 11.84GPSNGGGQQQRR186 pKa = 11.84SRR188 pKa = 11.84SRR190 pKa = 11.84VRR192 pKa = 11.84LNRR195 pKa = 11.84NNFNNNNVNNNNVNNRR211 pKa = 11.84NVNNRR216 pKa = 11.84NVNNRR221 pKa = 11.84NVNNQGSVRR230 pKa = 11.84RR231 pKa = 11.84SNLLKK236 pKa = 10.51SDD238 pKa = 4.13LNDD241 pKa = 3.36GFALLNQLRR250 pKa = 11.84RR251 pKa = 11.84FALNLPKK258 pKa = 10.47NRR260 pKa = 11.84SNQLNRR266 pKa = 11.84PLRR269 pKa = 11.84GRR271 pKa = 11.84IVKK274 pKa = 10.29RR275 pKa = 11.84NQNRR279 pKa = 11.84QTGNGAGVGGGRR291 pKa = 11.84IARR294 pKa = 11.84NIRR297 pKa = 11.84QNPNMADD304 pKa = 3.64GNRR307 pKa = 11.84QRR309 pKa = 11.84GKK311 pKa = 9.94IIKK314 pKa = 9.99RR315 pKa = 11.84SAQAQTRR322 pKa = 11.84GRR324 pKa = 11.84AVNRR328 pKa = 11.84KK329 pKa = 7.61QANQRR334 pKa = 11.84NGQGQAGRR342 pKa = 11.84RR343 pKa = 11.84GRR345 pKa = 11.84SRR347 pKa = 11.84TRR349 pKa = 11.84RR350 pKa = 11.84GVSNNNNNSRR360 pKa = 11.84GIIQNLKK367 pKa = 9.09QGHH370 pKa = 5.77KK371 pKa = 10.06VVEE374 pKa = 4.46VSS376 pKa = 3.19
MM1 pKa = 6.72NTSTRR6 pKa = 11.84LTLNDD11 pKa = 3.74RR12 pKa = 11.84FSMIKK17 pKa = 9.67QGNGVLPKK25 pKa = 8.84QQRR28 pKa = 11.84GRR30 pKa = 11.84SRR32 pKa = 11.84SRR34 pKa = 11.84SQTRR38 pKa = 11.84RR39 pKa = 11.84PSRR42 pKa = 11.84QLSQGGPRR50 pKa = 11.84KK51 pKa = 9.6NRR53 pKa = 11.84QLLDD57 pKa = 3.52QLEE60 pKa = 4.6KK61 pKa = 8.35QHH63 pKa = 6.03KK64 pKa = 8.75MRR66 pKa = 11.84MAMKK70 pKa = 10.05LKK72 pKa = 9.92NKK74 pKa = 10.23SIMRR78 pKa = 11.84SRR80 pKa = 11.84GRR82 pKa = 11.84VQSQNRR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84PANSLIATIKK100 pKa = 10.41RR101 pKa = 11.84KK102 pKa = 8.28MVEE105 pKa = 4.11DD106 pKa = 3.42SLKK109 pKa = 10.67RR110 pKa = 11.84GSNLTTFVPLNDD122 pKa = 3.51GPRR125 pKa = 11.84RR126 pKa = 11.84NFRR129 pKa = 11.84GRR131 pKa = 11.84SRR133 pKa = 11.84SRR135 pKa = 11.84SQSRR139 pKa = 11.84NRR141 pKa = 11.84NQQRR145 pKa = 11.84SQSRR149 pKa = 11.84NRR151 pKa = 11.84NQQQQQGSVIDD162 pKa = 3.83RR163 pKa = 11.84LGVRR167 pKa = 11.84QPLKK171 pKa = 10.34RR172 pKa = 11.84MRR174 pKa = 11.84RR175 pKa = 11.84GPSNGGGQQQRR186 pKa = 11.84SRR188 pKa = 11.84SRR190 pKa = 11.84VRR192 pKa = 11.84LNRR195 pKa = 11.84NNFNNNNVNNNNVNNRR211 pKa = 11.84NVNNRR216 pKa = 11.84NVNNRR221 pKa = 11.84NVNNQGSVRR230 pKa = 11.84RR231 pKa = 11.84SNLLKK236 pKa = 10.51SDD238 pKa = 4.13LNDD241 pKa = 3.36GFALLNQLRR250 pKa = 11.84RR251 pKa = 11.84FALNLPKK258 pKa = 10.47NRR260 pKa = 11.84SNQLNRR266 pKa = 11.84PLRR269 pKa = 11.84GRR271 pKa = 11.84IVKK274 pKa = 10.29RR275 pKa = 11.84NQNRR279 pKa = 11.84QTGNGAGVGGGRR291 pKa = 11.84IARR294 pKa = 11.84NIRR297 pKa = 11.84QNPNMADD304 pKa = 3.64GNRR307 pKa = 11.84QRR309 pKa = 11.84GKK311 pKa = 9.94IIKK314 pKa = 9.99RR315 pKa = 11.84SAQAQTRR322 pKa = 11.84GRR324 pKa = 11.84AVNRR328 pKa = 11.84KK329 pKa = 7.61QANQRR334 pKa = 11.84NGQGQAGRR342 pKa = 11.84RR343 pKa = 11.84GRR345 pKa = 11.84SRR347 pKa = 11.84TRR349 pKa = 11.84RR350 pKa = 11.84GVSNNNNNSRR360 pKa = 11.84GIIQNLKK367 pKa = 9.09QGHH370 pKa = 5.77KK371 pKa = 10.06VVEE374 pKa = 4.46VSS376 pKa = 3.19
Molecular weight: 43.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
8391700 |
14 |
15766 |
372.2 |
42.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.055 ± 0.014 | 1.982 ± 0.023 |
5.399 ± 0.016 | 6.783 ± 0.028 |
4.662 ± 0.021 | 4.677 ± 0.023 |
2.494 ± 0.011 | 6.716 ± 0.018 |
7.439 ± 0.026 | 8.97 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.496 ± 0.008 | 6.212 ± 0.019 |
4.334 ± 0.021 | 4.3 ± 0.019 |
4.781 ± 0.016 | 8.494 ± 0.03 |
5.634 ± 0.02 | 5.533 ± 0.014 |
0.951 ± 0.006 | 3.088 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
