
Schistosoma curassoni
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Platyhelminthes; Trematoda; Digenea; Strigeidida; Schistosomatoidea; Schistosomatidae; Schistosoma
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
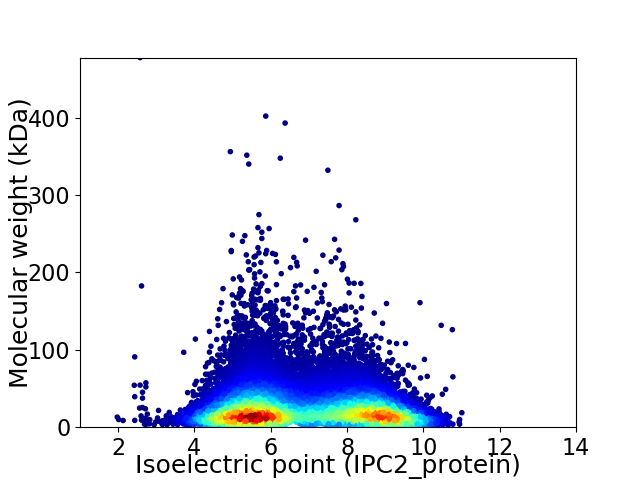
Virtual 2D-PAGE plot for 22688 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A183K9Y4|A0A183K9Y4_9TREM Uncharacterized protein OS=Schistosoma curassoni OX=6186 GN=SCUD_LOCUS11817 PE=4 SV=1
MM1 pKa = 7.28NLSLPQQFWTKK12 pKa = 10.73LFIVLNSCFVIFGIVLLALGIKK34 pKa = 10.24GLEE37 pKa = 4.1TLNKK41 pKa = 9.96FGTILTGIIPPIIPIAIFIGCLILIGTTIGYY72 pKa = 9.54IGLWKK77 pKa = 9.62PKK79 pKa = 10.16KK80 pKa = 9.69FIHH83 pKa = 6.45IACLCLAVIIEE94 pKa = 4.2IAIATATATMGEE106 pKa = 4.31KK107 pKa = 10.31FQTAANHH114 pKa = 6.08SVLQAVKK121 pKa = 10.14QFYY124 pKa = 10.54SNPYY128 pKa = 8.77CQVEE132 pKa = 4.01MDD134 pKa = 3.59KK135 pKa = 11.12LQRR138 pKa = 11.84NYY140 pKa = 9.79RR141 pKa = 11.84CCGATSYY148 pKa = 11.45LDD150 pKa = 3.52YY151 pKa = 11.22VQTNKK156 pKa = 9.09TVPFSCFIGTLVYY169 pKa = 10.5ARR171 pKa = 11.84HH172 pKa = 6.35IFDD175 pKa = 6.13DD176 pKa = 5.63DD177 pKa = 4.7DD178 pKa = 6.19ALDD181 pKa = 5.78DD182 pKa = 6.58DD183 pKa = 5.27DD184 pKa = 5.59TLDD187 pKa = 5.99DD188 pKa = 5.79DD189 pKa = 6.15ALDD192 pKa = 5.71DD193 pKa = 5.85DD194 pKa = 6.35DD195 pKa = 7.56DD196 pKa = 5.01DD197 pKa = 6.59ALDD200 pKa = 4.67EE201 pKa = 5.63DD202 pKa = 5.99DD203 pKa = 6.66DD204 pKa = 5.69DD205 pKa = 6.59ALDD208 pKa = 5.87DD209 pKa = 5.74DD210 pKa = 6.41DD211 pKa = 7.57DD212 pKa = 5.58DD213 pKa = 6.57ALDD216 pKa = 5.81DD217 pKa = 5.74DD218 pKa = 6.41DD219 pKa = 7.57DD220 pKa = 5.58DD221 pKa = 6.57ALDD224 pKa = 5.81DD225 pKa = 5.74DD226 pKa = 6.41DD227 pKa = 7.57DD228 pKa = 5.58DD229 pKa = 6.57ALDD232 pKa = 6.02DD233 pKa = 6.43DD234 pKa = 6.52DD235 pKa = 7.72DD236 pKa = 7.25DD237 pKa = 7.47DD238 pKa = 7.49DD239 pKa = 7.17DD240 pKa = 6.74GDD242 pKa = 4.16YY243 pKa = 11.47DD244 pKa = 5.31ALNDD248 pKa = 5.4DD249 pKa = 5.43DD250 pKa = 6.44ALDD253 pKa = 5.67DD254 pKa = 5.71DD255 pKa = 6.41DD256 pKa = 7.57DD257 pKa = 5.58DD258 pKa = 6.57ALDD261 pKa = 5.2DD262 pKa = 5.86DD263 pKa = 6.31DD264 pKa = 7.37DD265 pKa = 3.75ITDD268 pKa = 4.84FIRR271 pKa = 11.84KK272 pKa = 8.12FWQKK276 pKa = 10.6IFITLNTLFVIFNIILLILGIITQNTLSKK305 pKa = 10.04YY306 pKa = 5.68TTILNTPISAIIPTILFTGCFGFIAALIGYY336 pKa = 8.88IGLWKK341 pKa = 10.17PMNLIALMHH350 pKa = 6.95IISLCIVTLIEE361 pKa = 3.98IGIATTSAVMYY372 pKa = 9.7DD373 pKa = 3.19QFYY376 pKa = 7.35TTTHH380 pKa = 6.06SALLDD385 pKa = 3.71SVKK388 pKa = 10.29FYY390 pKa = 11.55YY391 pKa = 10.48EE392 pKa = 3.57NSQYY396 pKa = 10.82EE397 pKa = 4.2MEE399 pKa = 4.43MDD401 pKa = 3.59HH402 pKa = 6.69LQTEE406 pKa = 5.42FKK408 pKa = 10.84CCGAEE413 pKa = 4.02SYY415 pKa = 11.32LDD417 pKa = 3.69YY418 pKa = 11.04RR419 pKa = 11.84KK420 pKa = 10.52LGMNIPFSCIIGYY433 pKa = 9.13LVYY436 pKa = 10.88ARR438 pKa = 11.84VPLEE442 pKa = 4.04EE443 pKa = 4.59PFHH446 pKa = 6.13GVSNRR451 pKa = 11.84KK452 pKa = 9.63VITALIPLTAFKK464 pKa = 9.78ITDD467 pKa = 3.52SEE469 pKa = 4.32FFYY472 pKa = 11.35SDD474 pKa = 4.84LEE476 pKa = 5.17DD477 pKa = 4.05IMINKK482 pKa = 8.97
MM1 pKa = 7.28NLSLPQQFWTKK12 pKa = 10.73LFIVLNSCFVIFGIVLLALGIKK34 pKa = 10.24GLEE37 pKa = 4.1TLNKK41 pKa = 9.96FGTILTGIIPPIIPIAIFIGCLILIGTTIGYY72 pKa = 9.54IGLWKK77 pKa = 9.62PKK79 pKa = 10.16KK80 pKa = 9.69FIHH83 pKa = 6.45IACLCLAVIIEE94 pKa = 4.2IAIATATATMGEE106 pKa = 4.31KK107 pKa = 10.31FQTAANHH114 pKa = 6.08SVLQAVKK121 pKa = 10.14QFYY124 pKa = 10.54SNPYY128 pKa = 8.77CQVEE132 pKa = 4.01MDD134 pKa = 3.59KK135 pKa = 11.12LQRR138 pKa = 11.84NYY140 pKa = 9.79RR141 pKa = 11.84CCGATSYY148 pKa = 11.45LDD150 pKa = 3.52YY151 pKa = 11.22VQTNKK156 pKa = 9.09TVPFSCFIGTLVYY169 pKa = 10.5ARR171 pKa = 11.84HH172 pKa = 6.35IFDD175 pKa = 6.13DD176 pKa = 5.63DD177 pKa = 4.7DD178 pKa = 6.19ALDD181 pKa = 5.78DD182 pKa = 6.58DD183 pKa = 5.27DD184 pKa = 5.59TLDD187 pKa = 5.99DD188 pKa = 5.79DD189 pKa = 6.15ALDD192 pKa = 5.71DD193 pKa = 5.85DD194 pKa = 6.35DD195 pKa = 7.56DD196 pKa = 5.01DD197 pKa = 6.59ALDD200 pKa = 4.67EE201 pKa = 5.63DD202 pKa = 5.99DD203 pKa = 6.66DD204 pKa = 5.69DD205 pKa = 6.59ALDD208 pKa = 5.87DD209 pKa = 5.74DD210 pKa = 6.41DD211 pKa = 7.57DD212 pKa = 5.58DD213 pKa = 6.57ALDD216 pKa = 5.81DD217 pKa = 5.74DD218 pKa = 6.41DD219 pKa = 7.57DD220 pKa = 5.58DD221 pKa = 6.57ALDD224 pKa = 5.81DD225 pKa = 5.74DD226 pKa = 6.41DD227 pKa = 7.57DD228 pKa = 5.58DD229 pKa = 6.57ALDD232 pKa = 6.02DD233 pKa = 6.43DD234 pKa = 6.52DD235 pKa = 7.72DD236 pKa = 7.25DD237 pKa = 7.47DD238 pKa = 7.49DD239 pKa = 7.17DD240 pKa = 6.74GDD242 pKa = 4.16YY243 pKa = 11.47DD244 pKa = 5.31ALNDD248 pKa = 5.4DD249 pKa = 5.43DD250 pKa = 6.44ALDD253 pKa = 5.67DD254 pKa = 5.71DD255 pKa = 6.41DD256 pKa = 7.57DD257 pKa = 5.58DD258 pKa = 6.57ALDD261 pKa = 5.2DD262 pKa = 5.86DD263 pKa = 6.31DD264 pKa = 7.37DD265 pKa = 3.75ITDD268 pKa = 4.84FIRR271 pKa = 11.84KK272 pKa = 8.12FWQKK276 pKa = 10.6IFITLNTLFVIFNIILLILGIITQNTLSKK305 pKa = 10.04YY306 pKa = 5.68TTILNTPISAIIPTILFTGCFGFIAALIGYY336 pKa = 8.88IGLWKK341 pKa = 10.17PMNLIALMHH350 pKa = 6.95IISLCIVTLIEE361 pKa = 3.98IGIATTSAVMYY372 pKa = 9.7DD373 pKa = 3.19QFYY376 pKa = 7.35TTTHH380 pKa = 6.06SALLDD385 pKa = 3.71SVKK388 pKa = 10.29FYY390 pKa = 11.55YY391 pKa = 10.48EE392 pKa = 3.57NSQYY396 pKa = 10.82EE397 pKa = 4.2MEE399 pKa = 4.43MDD401 pKa = 3.59HH402 pKa = 6.69LQTEE406 pKa = 5.42FKK408 pKa = 10.84CCGAEE413 pKa = 4.02SYY415 pKa = 11.32LDD417 pKa = 3.69YY418 pKa = 11.04RR419 pKa = 11.84KK420 pKa = 10.52LGMNIPFSCIIGYY433 pKa = 9.13LVYY436 pKa = 10.88ARR438 pKa = 11.84VPLEE442 pKa = 4.04EE443 pKa = 4.59PFHH446 pKa = 6.13GVSNRR451 pKa = 11.84KK452 pKa = 9.63VITALIPLTAFKK464 pKa = 9.78ITDD467 pKa = 3.52SEE469 pKa = 4.32FFYY472 pKa = 11.35SDD474 pKa = 4.84LEE476 pKa = 5.17DD477 pKa = 4.05IMINKK482 pKa = 8.97
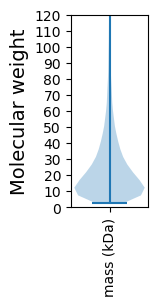
Molecular weight: 54.13 kDa
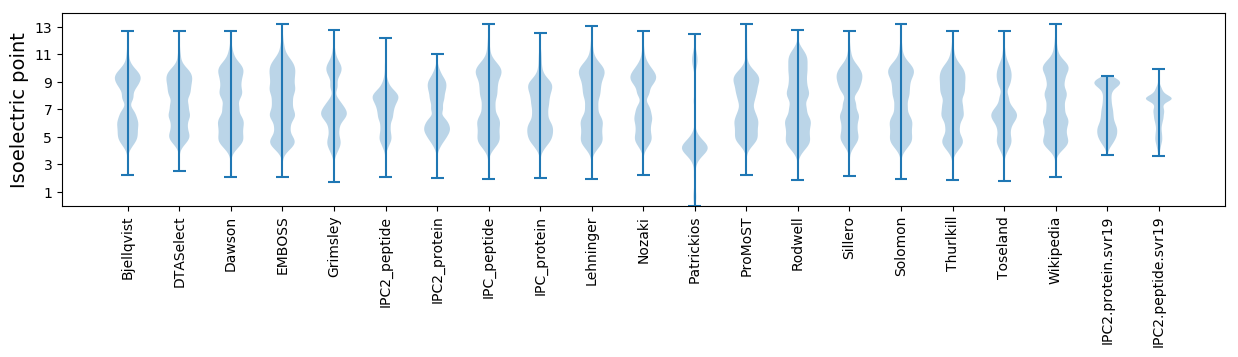
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A183JGX8|A0A183JGX8_9TREM Uncharacterized protein OS=Schistosoma curassoni OX=6186 GN=SCUD_LOCUS1952 PE=4 SV=1
MM1 pKa = 7.33MLTTKK6 pKa = 10.19RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84IRR11 pKa = 11.84TVKK14 pKa = 9.14RR15 pKa = 11.84TNPINAFQKK24 pKa = 9.18MRR26 pKa = 11.84TIINIII32 pKa = 3.49
MM1 pKa = 7.33MLTTKK6 pKa = 10.19RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84IRR11 pKa = 11.84TVKK14 pKa = 9.14RR15 pKa = 11.84TNPINAFQKK24 pKa = 9.18MRR26 pKa = 11.84TIINIII32 pKa = 3.49
Molecular weight: 3.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5552202 |
29 |
4267 |
244.7 |
27.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.506 ± 0.018 | 2.092 ± 0.012 |
5.36 ± 0.046 | 5.507 ± 0.021 |
3.799 ± 0.014 | 4.39 ± 0.024 |
2.921 ± 0.012 | 6.691 ± 0.02 |
5.769 ± 0.021 | 9.558 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.166 ± 0.015 | 7.165 ± 0.028 |
4.457 ± 0.017 | 4.326 ± 0.016 |
4.947 ± 0.017 | 9.985 ± 0.032 |
6.635 ± 0.038 | 5.359 ± 0.025 |
1.021 ± 0.006 | 3.23 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
