
California sea lion adenovirus 1
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Mastadenovirus; Sea lion mastadenovirus A
Average proteome isoelectric point is 4.43
Get precalculated fractions of proteins
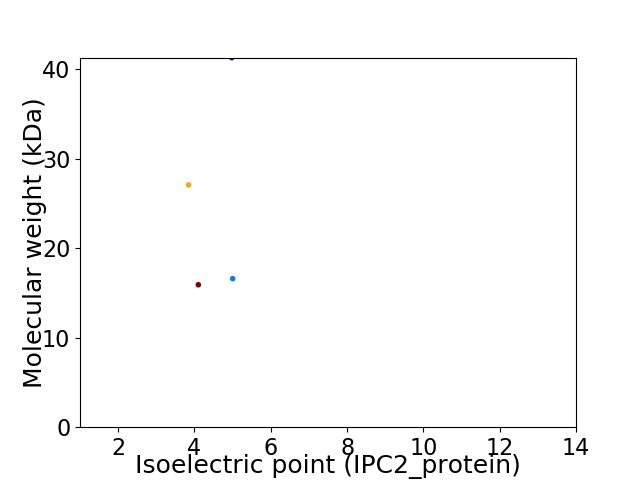
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
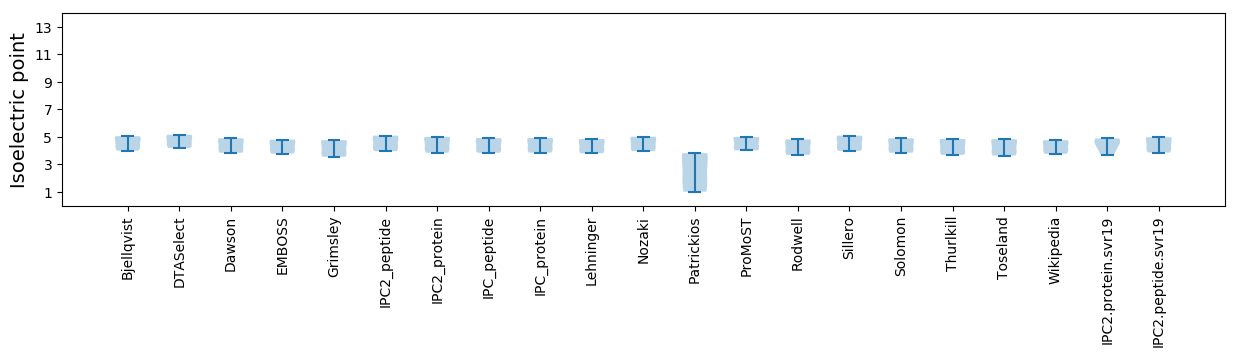
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7D5NTE2|A0A7D5NTE2_9ADEN IX OS=California sea lion adenovirus 1 OX=943083 PE=3 SV=1
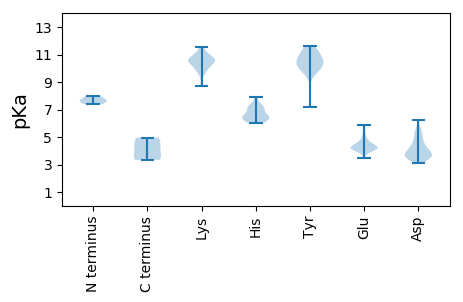
MM1 pKa = 7.67PLITLHH7 pKa = 6.5FSKK10 pKa = 10.74EE11 pKa = 3.95LLEE14 pKa = 4.19MVANCLPEE22 pKa = 4.56DD23 pKa = 3.85ARR25 pKa = 11.84PPHH28 pKa = 7.01PYY30 pKa = 10.61DD31 pKa = 4.72LLDD34 pKa = 4.86LEE36 pKa = 5.1HH37 pKa = 6.6QPSLHH42 pKa = 7.02DD43 pKa = 3.57MFDD46 pKa = 3.12IVLDD50 pKa = 3.76SDD52 pKa = 4.13VGGEE56 pKa = 4.14TPEE59 pKa = 4.62IEE61 pKa = 4.57LDD63 pKa = 3.8FPEE66 pKa = 4.47TLVNDD71 pKa = 3.92ALDD74 pKa = 3.96YY75 pKa = 10.17FASSPISIPSDD86 pKa = 3.5SSPVSPDD93 pKa = 3.52LCCHH97 pKa = 6.61EE98 pKa = 5.09SPLSSVSEE106 pKa = 4.29AEE108 pKa = 4.23NEE110 pKa = 4.03DD111 pKa = 3.59TSVEE115 pKa = 4.66KK116 pKa = 10.81EE117 pKa = 4.04NQDD120 pKa = 4.2FKK122 pKa = 11.37LDD124 pKa = 3.75CPDD127 pKa = 3.58YY128 pKa = 11.06PGVNCTSCDD137 pKa = 3.43YY138 pKa = 10.81HH139 pKa = 7.89RR140 pKa = 11.84QANPEE145 pKa = 4.17AVCSLCYY152 pKa = 9.87MRR154 pKa = 11.84ATSHH158 pKa = 7.22LIFEE162 pKa = 4.88PVSPPPVSEE171 pKa = 4.68DD172 pKa = 3.35VFDD175 pKa = 5.22FDD177 pKa = 5.14VDD179 pKa = 4.0TVLDD183 pKa = 4.01DD184 pKa = 6.22LIAEE188 pKa = 4.61KK189 pKa = 11.01DD190 pKa = 3.4PFLSFEE196 pKa = 4.38EE197 pKa = 4.16VDD199 pKa = 5.46ASLQCLANGANSLFTLDD216 pKa = 3.89SAPASTLKK224 pKa = 10.52RR225 pKa = 11.84KK226 pKa = 9.79HH227 pKa = 6.98DD228 pKa = 5.03DD229 pKa = 3.79DD230 pKa = 5.56DD231 pKa = 3.95QLEE234 pKa = 4.36PLDD237 pKa = 4.43LSLPKK242 pKa = 10.23KK243 pKa = 10.53SCC245 pKa = 3.34
MM1 pKa = 7.67PLITLHH7 pKa = 6.5FSKK10 pKa = 10.74EE11 pKa = 3.95LLEE14 pKa = 4.19MVANCLPEE22 pKa = 4.56DD23 pKa = 3.85ARR25 pKa = 11.84PPHH28 pKa = 7.01PYY30 pKa = 10.61DD31 pKa = 4.72LLDD34 pKa = 4.86LEE36 pKa = 5.1HH37 pKa = 6.6QPSLHH42 pKa = 7.02DD43 pKa = 3.57MFDD46 pKa = 3.12IVLDD50 pKa = 3.76SDD52 pKa = 4.13VGGEE56 pKa = 4.14TPEE59 pKa = 4.62IEE61 pKa = 4.57LDD63 pKa = 3.8FPEE66 pKa = 4.47TLVNDD71 pKa = 3.92ALDD74 pKa = 3.96YY75 pKa = 10.17FASSPISIPSDD86 pKa = 3.5SSPVSPDD93 pKa = 3.52LCCHH97 pKa = 6.61EE98 pKa = 5.09SPLSSVSEE106 pKa = 4.29AEE108 pKa = 4.23NEE110 pKa = 4.03DD111 pKa = 3.59TSVEE115 pKa = 4.66KK116 pKa = 10.81EE117 pKa = 4.04NQDD120 pKa = 4.2FKK122 pKa = 11.37LDD124 pKa = 3.75CPDD127 pKa = 3.58YY128 pKa = 11.06PGVNCTSCDD137 pKa = 3.43YY138 pKa = 10.81HH139 pKa = 7.89RR140 pKa = 11.84QANPEE145 pKa = 4.17AVCSLCYY152 pKa = 9.87MRR154 pKa = 11.84ATSHH158 pKa = 7.22LIFEE162 pKa = 4.88PVSPPPVSEE171 pKa = 4.68DD172 pKa = 3.35VFDD175 pKa = 5.22FDD177 pKa = 5.14VDD179 pKa = 4.0TVLDD183 pKa = 4.01DD184 pKa = 6.22LIAEE188 pKa = 4.61KK189 pKa = 11.01DD190 pKa = 3.4PFLSFEE196 pKa = 4.38EE197 pKa = 4.16VDD199 pKa = 5.46ASLQCLANGANSLFTLDD216 pKa = 3.89SAPASTLKK224 pKa = 10.52RR225 pKa = 11.84KK226 pKa = 9.79HH227 pKa = 6.98DD228 pKa = 5.03DD229 pKa = 3.79DD230 pKa = 5.56DD231 pKa = 3.95QLEE234 pKa = 4.36PLDD237 pKa = 4.43LSLPKK242 pKa = 10.23KK243 pKa = 10.53SCC245 pKa = 3.34
Molecular weight: 27.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A059XN88|A0A059XN88_9ADEN E1A OS=California sea lion adenovirus 1 OX=943083 PE=4 SV=1
MM1 pKa = 7.39EE2 pKa = 4.25QLKK5 pKa = 10.59DD6 pKa = 3.34YY7 pKa = 11.33SFFKK11 pKa = 10.69EE12 pKa = 4.46VVEE15 pKa = 4.23HH16 pKa = 6.98ASLRR20 pKa = 11.84TGNLYY25 pKa = 10.61RR26 pKa = 11.84FFFSSSLAKK35 pKa = 10.56VVFQVYY41 pKa = 9.71LEE43 pKa = 4.22KK44 pKa = 11.04VGEE47 pKa = 3.95FDD49 pKa = 5.36RR50 pKa = 11.84LDD52 pKa = 3.63STASALLALSQGNNFLFEE70 pKa = 4.14KK71 pKa = 10.66LIIPEE76 pKa = 4.13LPFYY80 pKa = 11.23SFGHH84 pKa = 6.11ILASLAFCSFCVNKK98 pKa = 10.38ADD100 pKa = 3.76ISATVSSGFILDD112 pKa = 4.17AVCRR116 pKa = 11.84PAWQKK121 pKa = 11.29FMMSHH126 pKa = 6.5NMMEE130 pKa = 4.25NQTGSSEE137 pKa = 4.14FSLEE141 pKa = 4.29EE142 pKa = 4.11ALTDD146 pKa = 3.58NN147 pKa = 4.96
MM1 pKa = 7.39EE2 pKa = 4.25QLKK5 pKa = 10.59DD6 pKa = 3.34YY7 pKa = 11.33SFFKK11 pKa = 10.69EE12 pKa = 4.46VVEE15 pKa = 4.23HH16 pKa = 6.98ASLRR20 pKa = 11.84TGNLYY25 pKa = 10.61RR26 pKa = 11.84FFFSSSLAKK35 pKa = 10.56VVFQVYY41 pKa = 9.71LEE43 pKa = 4.22KK44 pKa = 11.04VGEE47 pKa = 3.95FDD49 pKa = 5.36RR50 pKa = 11.84LDD52 pKa = 3.63STASALLALSQGNNFLFEE70 pKa = 4.14KK71 pKa = 10.66LIIPEE76 pKa = 4.13LPFYY80 pKa = 11.23SFGHH84 pKa = 6.11ILASLAFCSFCVNKK98 pKa = 10.38ADD100 pKa = 3.76ISATVSSGFILDD112 pKa = 4.17AVCRR116 pKa = 11.84PAWQKK121 pKa = 11.29FMMSHH126 pKa = 6.5NMMEE130 pKa = 4.25NQTGSSEE137 pKa = 4.14FSLEE141 pKa = 4.29EE142 pKa = 4.11ALTDD146 pKa = 3.58NN147 pKa = 4.96
Molecular weight: 16.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
966 |
147 |
418 |
241.5 |
25.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.797 ± 0.684 | 3.623 ± 0.951 |
7.557 ± 1.938 | 7.35 ± 0.389 |
4.969 ± 1.269 | 3.727 ± 0.701 |
2.484 ± 0.261 | 4.348 ± 0.803 |
4.037 ± 0.726 | 9.006 ± 1.384 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.656 ± 0.359 | 4.762 ± 0.629 |
5.383 ± 1.433 | 3.106 ± 1.368 |
2.692 ± 0.407 | 9.317 ± 1.003 |
4.141 ± 0.241 | 6.936 ± 0.703 |
0.518 ± 0.163 | 2.484 ± 0.35 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
