
Camel associated drosmacovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Drosmacovirus
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
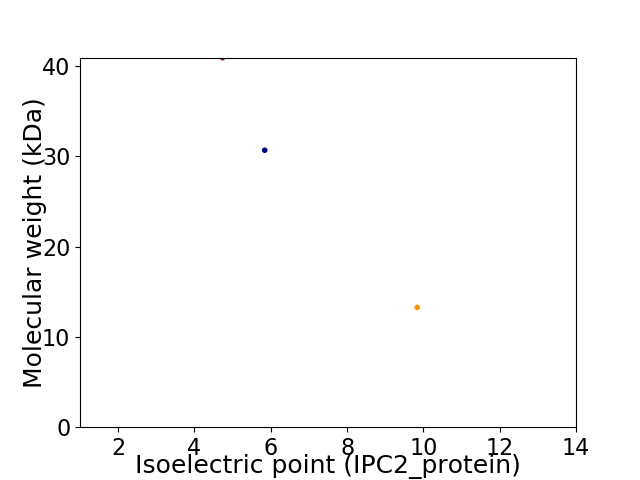
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1EKU7|A0A0A1EKU7_9VIRU Uncharacterized protein OS=Camel associated drosmacovirus 1 OX=2169876 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.98DD2 pKa = 4.79IEE4 pKa = 5.05GLTDD8 pKa = 3.75PLGVSDD14 pKa = 3.94TMGAEE19 pKa = 4.08GGEE22 pKa = 4.21MVTVSIEE29 pKa = 3.86EE30 pKa = 4.34MYY32 pKa = 11.77DD33 pKa = 3.02MATSLNKK40 pKa = 9.87IGIIGIHH47 pKa = 5.67TPNLGQIARR56 pKa = 11.84RR57 pKa = 11.84WAGLMINHH65 pKa = 7.1KK66 pKa = 8.62FLRR69 pKa = 11.84VKK71 pKa = 10.62SCDD74 pKa = 3.21VHH76 pKa = 6.4LACASMLPADD86 pKa = 4.53PLQIGTTTGTIAPQDD101 pKa = 3.55MMNPLLYY108 pKa = 10.0RR109 pKa = 11.84AVSNEE114 pKa = 3.16SWNAFTSRR122 pKa = 11.84LFATSGASVNQNSVKK137 pKa = 10.34VFTEE141 pKa = 4.13EE142 pKa = 4.99FSTQSDD148 pKa = 3.75EE149 pKa = 4.36NNEE152 pKa = 4.1KK153 pKa = 10.05IYY155 pKa = 11.2YY156 pKa = 9.13NALSEE161 pKa = 4.56PGWKK165 pKa = 9.73KK166 pKa = 10.69AFPQSGLDD174 pKa = 3.5INGLKK179 pKa = 10.18PLVYY183 pKa = 10.04PILTQFGNGEE193 pKa = 3.81AGINNFGLLGSSVQATTPAGNPTTVTNPTTGTAQLDD229 pKa = 3.36NARR232 pKa = 11.84YY233 pKa = 9.06FRR235 pKa = 11.84GKK237 pKa = 8.9AQPMPRR243 pKa = 11.84FPTSYY248 pKa = 9.83PYY250 pKa = 11.22VSTDD254 pKa = 2.82TGSTVGDD261 pKa = 3.27TWTLMFQDD269 pKa = 4.76SYY271 pKa = 11.29QIPRR275 pKa = 11.84TYY277 pKa = 9.88VACILTPPGKK287 pKa = 10.2LIKK290 pKa = 10.22FYY292 pKa = 10.93YY293 pKa = 9.61RR294 pKa = 11.84LRR296 pKa = 11.84IVWYY300 pKa = 9.84IEE302 pKa = 3.7FSDD305 pKa = 3.59MCTATEE311 pKa = 3.83RR312 pKa = 11.84VVGNSMVTAGNYY324 pKa = 9.8AYY326 pKa = 10.41SRR328 pKa = 11.84QYY330 pKa = 9.91TIPEE334 pKa = 4.14AVSSKK339 pKa = 9.5LTEE342 pKa = 4.71DD343 pKa = 3.38STLVTDD349 pKa = 3.95SDD351 pKa = 4.81LVSEE355 pKa = 5.33DD356 pKa = 4.52DD357 pKa = 4.73DD358 pKa = 4.71NSATAVNMVLDD369 pKa = 3.9KK370 pKa = 11.56VMEE373 pKa = 4.22KK374 pKa = 10.84
MM1 pKa = 7.98DD2 pKa = 4.79IEE4 pKa = 5.05GLTDD8 pKa = 3.75PLGVSDD14 pKa = 3.94TMGAEE19 pKa = 4.08GGEE22 pKa = 4.21MVTVSIEE29 pKa = 3.86EE30 pKa = 4.34MYY32 pKa = 11.77DD33 pKa = 3.02MATSLNKK40 pKa = 9.87IGIIGIHH47 pKa = 5.67TPNLGQIARR56 pKa = 11.84RR57 pKa = 11.84WAGLMINHH65 pKa = 7.1KK66 pKa = 8.62FLRR69 pKa = 11.84VKK71 pKa = 10.62SCDD74 pKa = 3.21VHH76 pKa = 6.4LACASMLPADD86 pKa = 4.53PLQIGTTTGTIAPQDD101 pKa = 3.55MMNPLLYY108 pKa = 10.0RR109 pKa = 11.84AVSNEE114 pKa = 3.16SWNAFTSRR122 pKa = 11.84LFATSGASVNQNSVKK137 pKa = 10.34VFTEE141 pKa = 4.13EE142 pKa = 4.99FSTQSDD148 pKa = 3.75EE149 pKa = 4.36NNEE152 pKa = 4.1KK153 pKa = 10.05IYY155 pKa = 11.2YY156 pKa = 9.13NALSEE161 pKa = 4.56PGWKK165 pKa = 9.73KK166 pKa = 10.69AFPQSGLDD174 pKa = 3.5INGLKK179 pKa = 10.18PLVYY183 pKa = 10.04PILTQFGNGEE193 pKa = 3.81AGINNFGLLGSSVQATTPAGNPTTVTNPTTGTAQLDD229 pKa = 3.36NARR232 pKa = 11.84YY233 pKa = 9.06FRR235 pKa = 11.84GKK237 pKa = 8.9AQPMPRR243 pKa = 11.84FPTSYY248 pKa = 9.83PYY250 pKa = 11.22VSTDD254 pKa = 2.82TGSTVGDD261 pKa = 3.27TWTLMFQDD269 pKa = 4.76SYY271 pKa = 11.29QIPRR275 pKa = 11.84TYY277 pKa = 9.88VACILTPPGKK287 pKa = 10.2LIKK290 pKa = 10.22FYY292 pKa = 10.93YY293 pKa = 9.61RR294 pKa = 11.84LRR296 pKa = 11.84IVWYY300 pKa = 9.84IEE302 pKa = 3.7FSDD305 pKa = 3.59MCTATEE311 pKa = 3.83RR312 pKa = 11.84VVGNSMVTAGNYY324 pKa = 9.8AYY326 pKa = 10.41SRR328 pKa = 11.84QYY330 pKa = 9.91TIPEE334 pKa = 4.14AVSSKK339 pKa = 9.5LTEE342 pKa = 4.71DD343 pKa = 3.38STLVTDD349 pKa = 3.95SDD351 pKa = 4.81LVSEE355 pKa = 5.33DD356 pKa = 4.52DD357 pKa = 4.73DD358 pKa = 4.71NSATAVNMVLDD369 pKa = 3.9KK370 pKa = 11.56VMEE373 pKa = 4.22KK374 pKa = 10.84
Molecular weight: 40.93 kDa
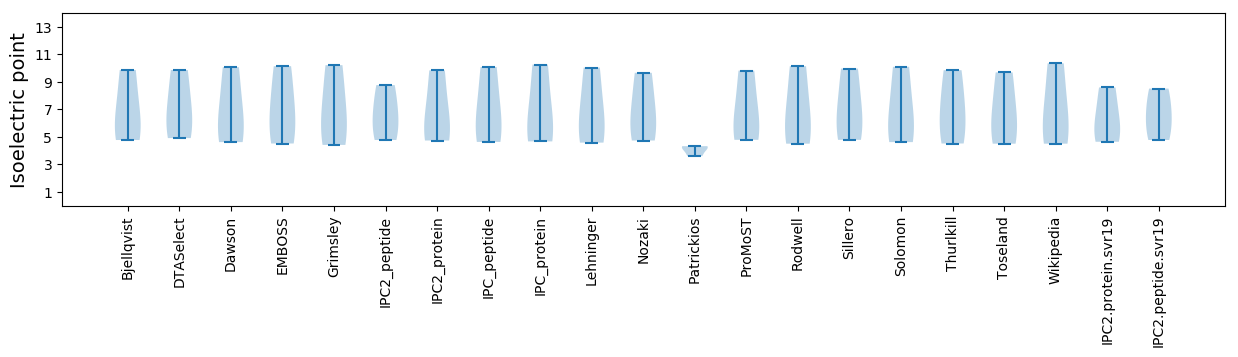
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1EL55|A0A0A1EL55_9VIRU Putative replicase protein OS=Camel associated drosmacovirus 1 OX=2169876 GN=rep PE=4 SV=1
MM1 pKa = 7.11SWSYY5 pKa = 10.86LYY7 pKa = 9.94WGQNVARR14 pKa = 11.84NSPSWTKK21 pKa = 11.18GNTNQWRR28 pKa = 11.84YY29 pKa = 10.06NMSGVPFVGDD39 pKa = 4.24WIRR42 pKa = 11.84AADD45 pKa = 3.7QNQWRR50 pKa = 11.84QDD52 pKa = 3.31YY53 pKa = 9.18MDD55 pKa = 4.0NYY57 pKa = 9.37GVGWEE62 pKa = 4.1DD63 pKa = 3.0VKK65 pKa = 11.26YY66 pKa = 10.46PSMVPGAGSYY76 pKa = 10.53AHH78 pKa = 7.29ASTNLIAGSGIVSKK92 pKa = 11.02NLLGLYY98 pKa = 10.2RR99 pKa = 11.84SGRR102 pKa = 11.84PYY104 pKa = 10.43KK105 pKa = 10.24AVPRR109 pKa = 11.84GGRR112 pKa = 11.84SVYY115 pKa = 10.56NRR117 pKa = 3.54
MM1 pKa = 7.11SWSYY5 pKa = 10.86LYY7 pKa = 9.94WGQNVARR14 pKa = 11.84NSPSWTKK21 pKa = 11.18GNTNQWRR28 pKa = 11.84YY29 pKa = 10.06NMSGVPFVGDD39 pKa = 4.24WIRR42 pKa = 11.84AADD45 pKa = 3.7QNQWRR50 pKa = 11.84QDD52 pKa = 3.31YY53 pKa = 9.18MDD55 pKa = 4.0NYY57 pKa = 9.37GVGWEE62 pKa = 4.1DD63 pKa = 3.0VKK65 pKa = 11.26YY66 pKa = 10.46PSMVPGAGSYY76 pKa = 10.53AHH78 pKa = 7.29ASTNLIAGSGIVSKK92 pKa = 11.02NLLGLYY98 pKa = 10.2RR99 pKa = 11.84SGRR102 pKa = 11.84PYY104 pKa = 10.43KK105 pKa = 10.24AVPRR109 pKa = 11.84GGRR112 pKa = 11.84SVYY115 pKa = 10.56NRR117 pKa = 3.54
Molecular weight: 13.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
752 |
117 |
374 |
250.7 |
28.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.984 ± 0.954 | 1.064 ± 0.218 |
5.718 ± 0.339 | 5.452 ± 1.229 |
3.191 ± 0.423 | 7.979 ± 0.722 |
1.33 ± 0.422 | 5.053 ± 0.451 |
5.319 ± 1.328 | 7.181 ± 0.531 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.59 ± 0.275 | 5.585 ± 0.735 |
5.452 ± 0.099 | 3.457 ± 0.175 |
4.787 ± 0.807 | 7.181 ± 1.051 |
7.314 ± 1.719 | 6.25 ± 0.517 |
3.059 ± 1.04 | 5.053 ± 0.656 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
