
Tortoise microvirus 92
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins
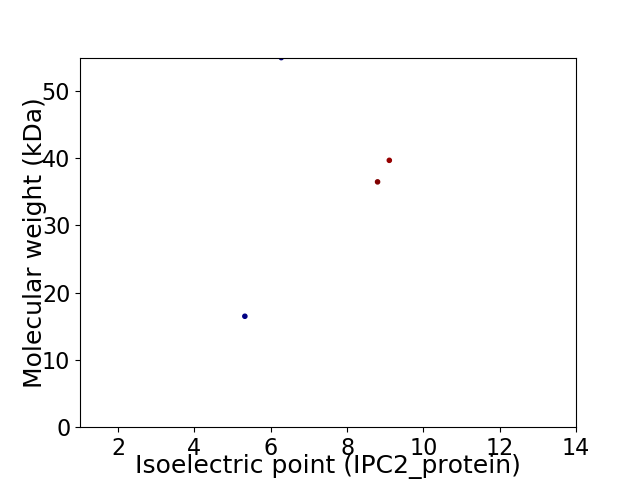
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
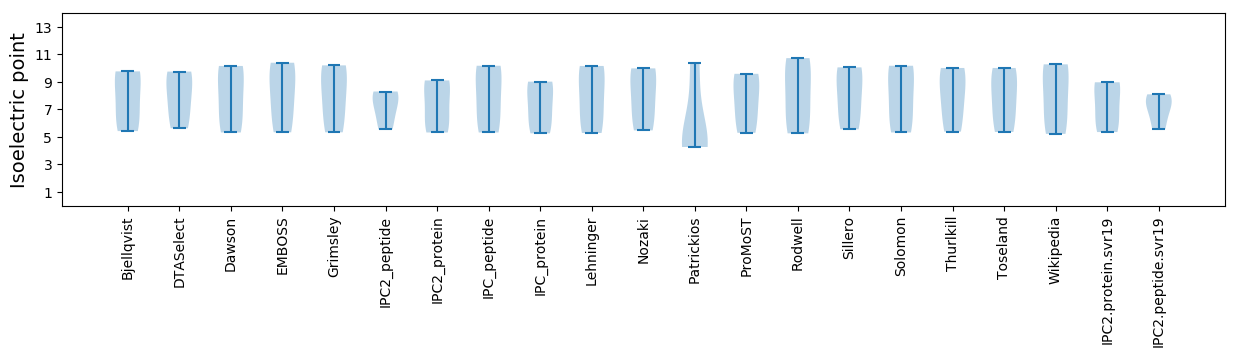
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8WAB6|A0A4P8WAB6_9VIRU Replication initiation protein OS=Tortoise microvirus 92 OX=2583201 PE=4 SV=1
MM1 pKa = 7.34AKK3 pKa = 10.07KK4 pKa = 8.47YY5 pKa = 10.46QKK7 pKa = 10.74YY8 pKa = 9.27EE9 pKa = 4.17EE10 pKa = 4.92PDD12 pKa = 3.61PTPVAVPVGFGHH24 pKa = 8.04PEE26 pKa = 3.66TLEE29 pKa = 3.83QKK31 pKa = 10.0IKK33 pKa = 10.65RR34 pKa = 11.84FVRR37 pKa = 11.84NEE39 pKa = 3.66RR40 pKa = 11.84YY41 pKa = 9.13QAALAAAGAEE51 pKa = 4.22TFEE54 pKa = 4.53EE55 pKa = 4.27SLDD58 pKa = 3.79FNVDD62 pKa = 3.53DD63 pKa = 6.68DD64 pKa = 4.74IDD66 pKa = 3.89PHH68 pKa = 7.12AHH70 pKa = 4.85STPWQEE76 pKa = 3.99RR77 pKa = 11.84ADD79 pKa = 3.85VEE81 pKa = 4.54TEE83 pKa = 3.64IEE85 pKa = 4.21DD86 pKa = 3.65SFRR89 pKa = 11.84AANSHH94 pKa = 6.86AEE96 pKa = 4.17MKK98 pKa = 10.63KK99 pKa = 10.51LEE101 pKa = 4.13ARR103 pKa = 11.84LKK105 pKa = 9.79EE106 pKa = 3.95LQKK109 pKa = 10.36IHH111 pKa = 6.57GPSEE115 pKa = 3.94PSSGGKK121 pKa = 9.17QNGRR125 pKa = 11.84PKK127 pKa = 10.66SKK129 pKa = 9.22STAPAKK135 pKa = 9.61TKK137 pKa = 10.35SGKK140 pKa = 10.17AGSDD144 pKa = 3.23ASDD147 pKa = 4.12DD148 pKa = 3.76ADD150 pKa = 3.28
MM1 pKa = 7.34AKK3 pKa = 10.07KK4 pKa = 8.47YY5 pKa = 10.46QKK7 pKa = 10.74YY8 pKa = 9.27EE9 pKa = 4.17EE10 pKa = 4.92PDD12 pKa = 3.61PTPVAVPVGFGHH24 pKa = 8.04PEE26 pKa = 3.66TLEE29 pKa = 3.83QKK31 pKa = 10.0IKK33 pKa = 10.65RR34 pKa = 11.84FVRR37 pKa = 11.84NEE39 pKa = 3.66RR40 pKa = 11.84YY41 pKa = 9.13QAALAAAGAEE51 pKa = 4.22TFEE54 pKa = 4.53EE55 pKa = 4.27SLDD58 pKa = 3.79FNVDD62 pKa = 3.53DD63 pKa = 6.68DD64 pKa = 4.74IDD66 pKa = 3.89PHH68 pKa = 7.12AHH70 pKa = 4.85STPWQEE76 pKa = 3.99RR77 pKa = 11.84ADD79 pKa = 3.85VEE81 pKa = 4.54TEE83 pKa = 3.64IEE85 pKa = 4.21DD86 pKa = 3.65SFRR89 pKa = 11.84AANSHH94 pKa = 6.86AEE96 pKa = 4.17MKK98 pKa = 10.63KK99 pKa = 10.51LEE101 pKa = 4.13ARR103 pKa = 11.84LKK105 pKa = 9.79EE106 pKa = 3.95LQKK109 pKa = 10.36IHH111 pKa = 6.57GPSEE115 pKa = 3.94PSSGGKK121 pKa = 9.17QNGRR125 pKa = 11.84PKK127 pKa = 10.66SKK129 pKa = 9.22STAPAKK135 pKa = 9.61TKK137 pKa = 10.35SGKK140 pKa = 10.17AGSDD144 pKa = 3.23ASDD147 pKa = 4.12DD148 pKa = 3.76ADD150 pKa = 3.28
Molecular weight: 16.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7W3|A0A4P8W7W3_9VIRU Uncharacterized protein OS=Tortoise microvirus 92 OX=2583201 PE=4 SV=1
MM1 pKa = 7.07EE2 pKa = 4.26TMLRR6 pKa = 11.84PGASTLLGIVCVTPSHH22 pKa = 7.24LLPKK26 pKa = 10.14KK27 pKa = 10.25NRR29 pKa = 11.84ALRR32 pKa = 11.84AQLPSNILVPEE43 pKa = 4.57FDD45 pKa = 3.44SKK47 pKa = 10.62IRR49 pKa = 11.84KK50 pKa = 9.07RR51 pKa = 11.84LPFALDD57 pKa = 3.08ANSARR62 pKa = 11.84KK63 pKa = 9.92SYY65 pKa = 10.3MLLKK69 pKa = 10.61KK70 pKa = 10.47LEE72 pKa = 4.02KK73 pKa = 10.39ALNVNALAYY82 pKa = 10.46QLTQKK87 pKa = 10.68SLAGGKK93 pKa = 8.79MDD95 pKa = 6.67PITAAAVISGGATIIGGLMNRR116 pKa = 11.84DD117 pKa = 3.57SQNSANAQNMEE128 pKa = 4.24INRR131 pKa = 11.84QNAEE135 pKa = 4.01LQKK138 pKa = 10.92EE139 pKa = 4.51FAQNGLRR146 pKa = 11.84WKK148 pKa = 10.91VSDD151 pKa = 3.52AKK153 pKa = 10.99AAGIHH158 pKa = 6.14PLAALGASTTSYY170 pKa = 11.07APAAIGVEE178 pKa = 4.65SNNAMGNAVAGMGQDD193 pKa = 2.81ISRR196 pKa = 11.84AVAATQTANEE206 pKa = 4.11RR207 pKa = 11.84EE208 pKa = 4.16MAEE211 pKa = 3.92MAKK214 pKa = 10.14QSLHH218 pKa = 7.08LDD220 pKa = 3.73LQGKK224 pKa = 9.09SLDD227 pKa = 3.55NAIRR231 pKa = 11.84ASQLAKK237 pKa = 10.89LNATQIGPPMPSAGTKK253 pKa = 9.21PVPSPQNPARR263 pKa = 11.84DD264 pKa = 3.67AGNITSYY271 pKa = 11.77AFMRR275 pKa = 11.84TDD277 pKa = 3.65DD278 pKa = 4.28GGLSVVPSEE287 pKa = 4.14AAKK290 pKa = 10.59EE291 pKa = 3.92RR292 pKa = 11.84TEE294 pKa = 4.75DD295 pKa = 5.02DD296 pKa = 3.86FVQQLGWAARR306 pKa = 11.84NQLVPAVSGLTPPSLKK322 pKa = 10.05EE323 pKa = 3.7HH324 pKa = 7.13PLPKK328 pKa = 9.23GQQWVWNPLKK338 pKa = 10.51QAFYY342 pKa = 10.06PSSGPSAVDD351 pKa = 2.91VYY353 pKa = 11.11NFRR356 pKa = 11.84KK357 pKa = 10.31GGPKK361 pKa = 9.03KK362 pKa = 9.82WKK364 pKa = 10.0IFNDD368 pKa = 3.67KK369 pKa = 10.44YY370 pKa = 11.42
MM1 pKa = 7.07EE2 pKa = 4.26TMLRR6 pKa = 11.84PGASTLLGIVCVTPSHH22 pKa = 7.24LLPKK26 pKa = 10.14KK27 pKa = 10.25NRR29 pKa = 11.84ALRR32 pKa = 11.84AQLPSNILVPEE43 pKa = 4.57FDD45 pKa = 3.44SKK47 pKa = 10.62IRR49 pKa = 11.84KK50 pKa = 9.07RR51 pKa = 11.84LPFALDD57 pKa = 3.08ANSARR62 pKa = 11.84KK63 pKa = 9.92SYY65 pKa = 10.3MLLKK69 pKa = 10.61KK70 pKa = 10.47LEE72 pKa = 4.02KK73 pKa = 10.39ALNVNALAYY82 pKa = 10.46QLTQKK87 pKa = 10.68SLAGGKK93 pKa = 8.79MDD95 pKa = 6.67PITAAAVISGGATIIGGLMNRR116 pKa = 11.84DD117 pKa = 3.57SQNSANAQNMEE128 pKa = 4.24INRR131 pKa = 11.84QNAEE135 pKa = 4.01LQKK138 pKa = 10.92EE139 pKa = 4.51FAQNGLRR146 pKa = 11.84WKK148 pKa = 10.91VSDD151 pKa = 3.52AKK153 pKa = 10.99AAGIHH158 pKa = 6.14PLAALGASTTSYY170 pKa = 11.07APAAIGVEE178 pKa = 4.65SNNAMGNAVAGMGQDD193 pKa = 2.81ISRR196 pKa = 11.84AVAATQTANEE206 pKa = 4.11RR207 pKa = 11.84EE208 pKa = 4.16MAEE211 pKa = 3.92MAKK214 pKa = 10.14QSLHH218 pKa = 7.08LDD220 pKa = 3.73LQGKK224 pKa = 9.09SLDD227 pKa = 3.55NAIRR231 pKa = 11.84ASQLAKK237 pKa = 10.89LNATQIGPPMPSAGTKK253 pKa = 9.21PVPSPQNPARR263 pKa = 11.84DD264 pKa = 3.67AGNITSYY271 pKa = 11.77AFMRR275 pKa = 11.84TDD277 pKa = 3.65DD278 pKa = 4.28GGLSVVPSEE287 pKa = 4.14AAKK290 pKa = 10.59EE291 pKa = 3.92RR292 pKa = 11.84TEE294 pKa = 4.75DD295 pKa = 5.02DD296 pKa = 3.86FVQQLGWAARR306 pKa = 11.84NQLVPAVSGLTPPSLKK322 pKa = 10.05EE323 pKa = 3.7HH324 pKa = 7.13PLPKK328 pKa = 9.23GQQWVWNPLKK338 pKa = 10.51QAFYY342 pKa = 10.06PSSGPSAVDD351 pKa = 2.91VYY353 pKa = 11.11NFRR356 pKa = 11.84KK357 pKa = 10.31GGPKK361 pKa = 9.03KK362 pKa = 9.82WKK364 pKa = 10.0IFNDD368 pKa = 3.67KK369 pKa = 10.44YY370 pKa = 11.42
Molecular weight: 39.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1329 |
150 |
492 |
332.3 |
36.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.481 ± 1.942 | 0.978 ± 0.705 |
5.568 ± 0.731 | 5.568 ± 1.094 |
3.386 ± 0.587 | 7.901 ± 0.53 |
2.408 ± 0.467 | 4.665 ± 0.508 |
6.622 ± 1.136 | 7.449 ± 0.865 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.558 ± 0.339 | 4.063 ± 0.906 |
5.944 ± 0.551 | 3.762 ± 0.728 |
6.396 ± 0.907 | 6.546 ± 0.572 |
6.471 ± 0.978 | 5.117 ± 0.686 |
1.354 ± 0.15 | 3.762 ± 0.864 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
